Identification, characterization and benefits of an exclusion system in an integrative and conjugative element of Bacillus subtilis
- PMID: 31361051
- PMCID: PMC6827876
- DOI: 10.1111/mmi.14359
Identification, characterization and benefits of an exclusion system in an integrative and conjugative element of Bacillus subtilis
Abstract
Integrative and conjugative elements (ICEs) are mobile genetic elements that transfer from cell to cell by conjugation (like plasmids) and integrate into the chromosomes of bacterial hosts (like lysogenic phages or transposons). ICEs are prevalent in bacterial chromosomes and play a major role in bacterial evolution by promoting horizontal gene transfer. Exclusion prevents the redundant transfer of conjugative elements into host cells that already contain a copy of the element. Exclusion has been characterized mostly for conjugative elements of Gram-negative bacteria. Here, we report the identification and characterization of an exclusion mechanism in ICEBs1 from the Gram-positive bacterium Bacillus subtilis. We found that cells containing ICEBs1 inhibit the activity of the ICEBs1-encoded conjugation machinery in other cells. This inhibition (exclusion) was specific to the cognate conjugation machinery and the ICEBs1 gene yddJ was both necessary and sufficient to mediate exclusion by recipient cells. Through a mutagenesis and enrichment screen, we identified exclusion-resistant mutations in the ICEBs1 gene conG. Using genes from a heterologous but related ICE, we found that the exclusion specificity was determined by ConG and YddJ. Finally, we found that under conditions that support conjugation, exclusion provides a selective advantage to the element and its host cells.
© 2019 John Wiley & Sons Ltd.
Conflict of interest statement
Figures

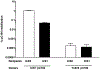
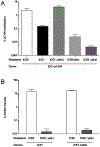



Similar articles
-
Specificity and Selective Advantage of an Exclusion System in the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.J Bacteriol. 2021 Apr 21;203(10):e00700-20. doi: 10.1128/JB.00700-20. Print 2021 Apr 21. J Bacteriol. 2021. PMID: 33649151 Free PMC article.
-
Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis.Plasmid. 2016 Jul;86:14-25. doi: 10.1016/j.plasmid.2016.07.001. Epub 2016 Jul 2. Plasmid. 2016. PMID: 27381852 Review.
-
Autonomous plasmid-like replication of a conjugative transposon.Mol Microbiol. 2010 Jan;75(2):268-79. doi: 10.1111/j.1365-2958.2009.06985.x. Epub 2009 Nov 25. Mol Microbiol. 2010. PMID: 19943900 Free PMC article.
-
Characterization of ConE, the VirB4 Homolog of the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.J Bacteriol. 2023 Jun 27;205(6):e0003323. doi: 10.1128/jb.00033-23. Epub 2023 May 23. J Bacteriol. 2023. PMID: 37219457 Free PMC article.
-
Integrative and Conjugative Elements (ICEs): What They Do and How They Work.Annu Rev Genet. 2015;49:577-601. doi: 10.1146/annurev-genet-112414-055018. Epub 2015 Oct 14. Annu Rev Genet. 2015. PMID: 26473380 Free PMC article. Review.
Cited by
-
Activation and modulation of the host response to DNA damage by an integrative and conjugative element.bioRxiv [Preprint]. 2024 Oct 9:2024.10.09.617469. doi: 10.1101/2024.10.09.617469. bioRxiv. 2024. Update in: J Bacteriol. 2025 Feb 20;207(2):e0046224. doi: 10.1128/jb.00462-24. PMID: 39416164 Free PMC article. Updated. Preprint.
-
The extended mobility of plasmids.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf652. doi: 10.1093/nar/gkaf652. Nucleic Acids Res. 2025. PMID: 40694848 Free PMC article. Review.
-
Contribution of the Mobilome to the Configuration of the Resistome of Corynebacterium striatum.Int J Mol Sci. 2024 Sep 29;25(19):10499. doi: 10.3390/ijms251910499. Int J Mol Sci. 2024. PMID: 39408827 Free PMC article.
-
Activation and modulation of the host response to DNA damage by an integrative and conjugative element.J Bacteriol. 2025 Feb 20;207(2):e0046224. doi: 10.1128/jb.00462-24. Epub 2025 Jan 23. J Bacteriol. 2025. PMID: 39846752 Free PMC article.
-
Second-Generation Transfer Mediates Efficient Propagation of ICEBs1 in Biofilms.J Bacteriol. 2022 Oct 18;204(10):e0018122. doi: 10.1128/jb.00181-22. Epub 2022 Sep 15. J Bacteriol. 2022. PMID: 36106856 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

