ULK2 is essential for degradation of ubiquitinated protein aggregates and homeostasis in skeletal muscle
- PMID: 31361156
- PMCID: PMC6902739
- DOI: 10.1096/fj.201900766R
ULK2 is essential for degradation of ubiquitinated protein aggregates and homeostasis in skeletal muscle
Abstract
Basal protein turnover, which largely relies on the degradation of ubiquitinated substrates, is instrumental for maintenance of muscle mass and function. However, the regulation of ubiquitinated protein degradation in healthy, nonatrophying skeletal muscle is still evolving, and potential tissue-specific modulators remain unknown. Using an unbiased expression analysis of 34 putative autophagy genes across mouse tissues, we identified unc-51 like autophagy activating kinase (Ulk)2, a homolog of the yeast autophagy related protein 1, as particularly enriched in skeletal muscle. Subsequent experiments revealed accumulations of insoluble ubiquitinated protein aggregates associated with the adaptors sequestosome 1 (SQSTM1, also known as p62) and next to breast cancer type 1 susceptibility protein gene 1 protein (NBR1) in adult muscles with ULK2 deficiency. ULK2 deficiency also led to impaired muscle force and caused myofiber atrophy and degeneration. These features were not observed in muscles with deficiency of the ULK2 paralog, ULK1. Furthermore, short-term ULK2 deficiency did not impair autophagy initiation, autophagosome to lysosome fusion, or protease activities of the lysosome and proteasome. Altogether, our results indicate that skeletal muscle ULK2 has a unique role in basal selective protein degradation by stimulating the recognition and proteolytic sequestration of insoluble ubiquitinated protein aggregates associated with p62 and NBR1. These findings have potential implications for conditions of poor protein homeostasis in muscles as observed in several myopathies and aging.-Fuqua, J. D., Mere, C. P., Kronemberger, A., Blomme, J., Bae, D., Turner, K. D., Harris, M. P., Scudese, E., Edwards, M., Ebert, S. M., de Sousa, L. G. O., Bodine, S. C., Yang, L., Adams, C. M., Lira, V. A. ULK2 is essential for degradation of ubiquitinated protein aggregates and homeostasis in skeletal muscle.
Keywords: NBR1; ULK1; aggrephagy; autophagy; p62; proteostasis.
Conflict of interest statement
The authors acknowledge use of the University of Iowa Central Microscopy Research Facility, a core resource supported by the University of Iowa Vice President for Research, and the Carver College of Medicine. This study was supported by a Fraternal Order of Eagles Pilot Research Grant and American Heart Association Grant 16SDG30360001 (to V.A.L.). The authors declare no conflicts of interest.
Figures

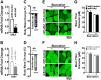
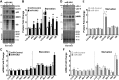
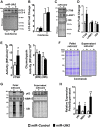
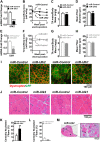
References
-
- Visser M., Schaap L. A. (2011) Consequences of sarcopenia. Clin. Geriatr. Med. 27, 387–399 - PubMed
-
- Kamel H. K. (2003) Sarcopenia and aging. Nutr. Rev. 61, 157–167 - PubMed
-
- Walrand S., Guillet C., Salles J., Cano N., Boirie Y. (2011) Physiopathological mechanism of sarcopenia. Clin. Geriatr. Med. 27, 365–385 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

