Genome-wide association analysis of 350 000 Caucasians from the UK Biobank identifies novel loci for asthma, hay fever and eczema
- PMID: 31361310
- PMCID: PMC6969355
- DOI: 10.1093/hmg/ddz175
Genome-wide association analysis of 350 000 Caucasians from the UK Biobank identifies novel loci for asthma, hay fever and eczema
Abstract
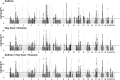
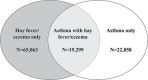
Even though heritability estimates suggest that the risk of asthma, hay fever and eczema is largely due to genetic factors, previous studies have not explained a large part of the genetics behind these diseases. In this genome-wide association study, we include 346 545 Caucasians from the UK Biobank to identify novel loci for asthma, hay fever and eczema and replicate novel loci in three independent cohorts. We further investigate if associated lead single nucleotide polymorphisms (SNPs) have a significantly larger effect for one disease compared to the other diseases, to highlight possible disease-specific effects. We identified 141 loci, of which 41 are novel, to be associated (P ≤ 3 × 10-8) with asthma, hay fever or eczema, analyzed separately or as disease phenotypes that includes the presence of different combinations of these diseases. The largest number of loci was associated with the combined phenotype (asthma/hay fever/eczema). However, as many as 20 loci had a significantly larger effect on hay fever/eczema only compared to their effects on asthma, while 26 loci exhibited larger effects on asthma compared with their effects on hay fever/eczema. At four of the novel loci, TNFRSF8, MYRF, TSPAN8, and BHMG1, the lead SNPs were in Linkage Disequilibrium (LD) (>0.8) with potentially casual missense variants. Our study shows that a large amount of the genetic contribution is shared between the diseases. Nonetheless, a number of SNPs have a significantly larger effect on one of the phenotypes, suggesting that part of the genetic contribution is more phenotype specific.
© The Author(s) 2019. Published by Oxford University Press.
Figures
References
-
- Ferreira M.A.R., Matheson M.C., Tang C.S., Granell R., Ang W., Hui J., Kiefer A.K., Duffy D.L., Baltic S., Danoy P. et al. (2014) Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J. Allergy Clin. Immunol., 133, 1564–1571. - PMC - PubMed
-
- Hinds D.A., McMahon G., Kiefer A.K., Do C.B., Eriksson N., Evans D.M., St Pourcain B., Ring S.M., Mountain J.L., Francke U. et al. (2013) A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet., 45, 907–911. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

