Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer
- PMID: 31361600
- PMCID: PMC6763249
- DOI: 10.1172/JCI128212
Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer
Abstract
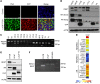
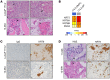
Metastatic castration-resistant prostate cancer (mCRPC) is a heterogeneous disease with diverse drivers of disease progression and mechanisms of therapeutic resistance. We conducted deep phenotypic characterization of CRPC metastases and patient-derived xenograft (PDX) lines using whole genome RNA sequencing, gene set enrichment analysis and immunohistochemistry. Our analyses revealed five mCRPC phenotypes based on the expression of well-characterized androgen receptor (AR) or neuroendocrine (NE) genes: (i) AR-high tumors (ARPC), (ii) AR-low tumors (ARLPC), (iii) amphicrine tumors composed of cells co-expressing AR and NE genes (AMPC), (iv) double-negative tumors (i.e. AR-/NE-; DNPC) and (v) tumors with small cell or NE gene expression without AR activity (SCNPC). RE1-silencing transcription factor (REST) activity, which suppresses NE gene expression, was lost in AMPC and SCNPC PDX models. However, knockdown of REST in cell lines revealed that attenuated REST activity drives the AMPC phenotype but is not sufficient for SCNPC conversion. We also identified a subtype of DNPC tumors with squamous differentiation and generated an encompassing 26-gene transcriptional signature that distinguished the five mCRPC phenotypes. Together, our data highlight the central role of AR and REST in classifying treatment-resistant mCRPC phenotypes. These molecular classifications could potentially guide future therapeutic studies and clinical trial design.
Keywords: Cell Biology; Expression profiling; Molecular pathology; Oncology; Prostate cancer.
Conflict of interest statement
Figures






Comment in
-
New classification may assist the development of targeted therapies for treatment-refractory castration-resistant prostate cancer.Transl Androl Urol. 2020 Apr;9(2):837-839. doi: 10.21037/tau.2020.03.24. Transl Androl Urol. 2020. PMID: 32420195 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

