Non-coding RNAs in cancers with chromosomal rearrangements: the signatures, causes, functions and implications
- PMID: 31361891
- PMCID: PMC6884712
- DOI: 10.1093/jmcb/mjz080
Non-coding RNAs in cancers with chromosomal rearrangements: the signatures, causes, functions and implications
Abstract
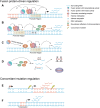
Chromosomal translocation leads to the juxtaposition of two otherwise separate DNA loci, which could result in gene fusion. These rearrangements at the DNA level are catastrophic events and often have causal roles in tumorigenesis. The oncogenic DNA messages are transferred to RNA molecules, which are in most cases translated into cancerous fusion proteins. Gene expression programs and signaling pathways are altered in these cytogenetically abnormal contexts. Notably, non-coding RNAs have attracted increasing attention and are believed to be tightly associated with chromosome-rearranged cancers. These RNAs not only function as modulators in downstream pathways but also directly affect chromosomal translocation or the associated products. This review summarizes recent research advances on the relationship between non-coding RNAs and chromosomal translocations and on diverse functions of non-coding RNAs in cancers with chromosomal rearrangements.
Keywords: chromosomal translocation; fusion protein; gene regulation; non-coding RNA; non-coding fusion transcript.
© The Author(s) (2019). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, IBCB, SIBS, CAS.
Figures


References
-
- Antonescu C.R., Agaram N.P., Sung Y.S., et al. (2018). A distinct malignant epithelioid neoplasm with GLI1 gene rearrangements, frequent S100 protein expression, and metastatic potential: expanding the spectrum of pathologic entities with ACTB/MALAT1/PTCH1–GLI1 fusions. Am. J. Surg. Pathol. 42, 553–560. - PMC - PubMed

