Rational Design of Artificial Metalloproteins and Metalloenzymes with Metal Clusters
- PMID: 31362341
- PMCID: PMC6696605
- DOI: 10.3390/molecules24152743
Rational Design of Artificial Metalloproteins and Metalloenzymes with Metal Clusters
Abstract
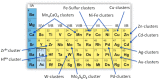
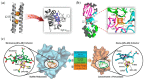
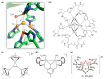
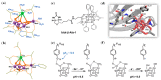
Metalloproteins and metalloenzymes play important roles in biological systems by using the limited metal ions, complexes, and clusters that are associated with the protein matrix. The design of artificial metalloproteins and metalloenzymes not only reveals the structure and function relationship of natural proteins, but also enables the synthesis of artificial proteins and enzymes with improved properties and functions. Acknowledging the progress in rational design from single to multiple active sites, this review focuses on recent achievements in the design of artificial metalloproteins and metalloenzymes with metal clusters, including zinc clusters, cadmium clusters, iron-sulfur clusters, and copper-sulfur clusters, as well as noble metal clusters and others. These metal clusters were designed in both native and de novo protein scaffolds for structural roles, electron transfer, or catalysis. Some synthetic metal clusters as functional models of native enzymes are also discussed. These achievements provide valuable insights for deep understanding of the natural proteins and enzymes, and practical clues for the further design of artificial enzymes with functions comparable or even beyond those of natural counterparts.
Keywords: metalclusters; metalloenzymes; metalloproteins; protein design; synthetic models.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Abiological catalysis by artificial haem proteins containing noble metals in place of iron.Nature. 2016 Jun 23;534(7608):534-7. doi: 10.1038/nature17968. Epub 2016 Jun 13. Nature. 2016. PMID: 27296224 Free PMC article.
-
Computational approaches for de novo design and redesign of metal-binding sites on proteins.Biosci Rep. 2017 Mar 27;37(2):BSR20160179. doi: 10.1042/BSR20160179. Print 2017 Apr 28. Biosci Rep. 2017. PMID: 28167677 Free PMC article. Review.
-
Design and engineering of artificial metalloproteins: from de novo metal coordination to catalysis.Protein Eng Des Sel. 2021 Feb 15;34:gzab003. doi: 10.1093/protein/gzab003. Protein Eng Des Sel. 2021. PMID: 33635315 Review.
-
Expansion of Redox Chemistry in Designer Metalloenzymes.Acc Chem Res. 2019 Mar 19;52(3):557-565. doi: 10.1021/acs.accounts.8b00627. Epub 2019 Feb 28. Acc Chem Res. 2019. PMID: 30816694 Review.
-
Repurposing metalloproteins as mimics of natural metalloenzymes for small-molecule activation.J Inorg Biochem. 2021 Jun;219:111430. doi: 10.1016/j.jinorgbio.2021.111430. Epub 2021 Mar 18. J Inorg Biochem. 2021. PMID: 33873051 Free PMC article. Review.
Cited by
-
Changing the tracks: screening for electron transfer proteins to support hydrogen production.J Biol Inorg Chem. 2022 Oct;27(7):631-640. doi: 10.1007/s00775-022-01956-1. Epub 2022 Aug 29. J Biol Inorg Chem. 2022. PMID: 36038787 Free PMC article.
-
Pyrrolyl and Indolyl α-γ-Diketo Acid Derivatives Acting as Selective Inhibitors of Human Carbonic Anhydrases IX and XII.Pharmaceuticals (Basel). 2023 Jan 27;16(2):188. doi: 10.3390/ph16020188. Pharmaceuticals (Basel). 2023. PMID: 37259337 Free PMC article.
-
Dye Decolorization by a Miniaturized Peroxidase Fe-MimochromeVI*a.Int J Mol Sci. 2023 Jul 4;24(13):11070. doi: 10.3390/ijms241311070. Int J Mol Sci. 2023. PMID: 37446248 Free PMC article.
-
Exploring the Reactivity of Polyoxometalates toward Proteins: From Interactions to Mechanistic Insights.JACS Au. 2023 Feb 13;3(4):978-990. doi: 10.1021/jacsau.3c00011. eCollection 2023 Apr 24. JACS Au. 2023. PMID: 37124292 Free PMC article. Review.
-
Comparative Analysis of Hulless Barley Transcriptomes to Regulatory Effects of Phosphorous Deficiency.Life (Basel). 2024 Jul 19;14(7):904. doi: 10.3390/life14070904. Life (Basel). 2024. PMID: 39063656 Free PMC article.
References
-
- Riordan J.F., Vallee B.L. The functional roles of metals in metalloenzymes. Adv. Exp. Med. Biol. 1974;48:33–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

