Relating evolutionary selection and mutant clonal dynamics in normal epithelia
- PMID: 31362624
- PMCID: PMC6685019
- DOI: 10.1098/rsif.2019.0230
Relating evolutionary selection and mutant clonal dynamics in normal epithelia
Abstract
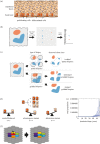
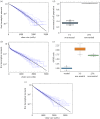
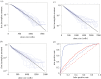
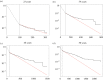
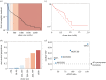
Cancer develops from mutated cells in normal tissues. Whether somatic mutations alter normal cell dynamics is key to understanding cancer risk and guiding interventions to reduce it. An analysis of the first incomplete moment of size distributions of clones carrying cancer-associated mutations in normal human eyelid skin gives a good fit with neutral drift, arguing mutations do not affect cell fate. However, this suggestion conflicts with genetic evidence in the same dataset that argues for strong positive selection of a subset of mutations. This implies cells carrying these mutations have a competitive advantage over normal cells, leading to large clonal expansions within the tissue. In the normal epithelium, clone growth is constrained by the limited size of the proliferating compartment and competition with surrounding cells. We show that if these factors are taken into account, the first incomplete moment of the clone size distribution is unable to exclude non-neutral behaviour. Furthermore, experimental factors can make a non-neutral clone size distribution appear neutral. We validate these principles with a new experimental dataset showing that when experiments are appropriately designed, the first incomplete moment can be a useful indicator of non-neutral competition. Finally, we discuss the complex relationship between mutant clone sizes and genetic selection.
Keywords: DNA sequencing; cancer; oesophagus; stem cells.
Conflict of interest statement
We declare we have no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

