Microbiota and gut ultrastructure of Anisakis pegreffii isolated from stranded cetaceans in the Adriatic Sea
- PMID: 31362767
- PMCID: PMC6668197
- DOI: 10.1186/s13071-019-3636-z
Microbiota and gut ultrastructure of Anisakis pegreffii isolated from stranded cetaceans in the Adriatic Sea
Abstract
Background: Inferring the microbiota diversity of helminths enables depiction of evolutionarily established ecological and pathological traits that characterize a particular parasite-host interaction. In turn, these traits could provide valuable information for the development of parasitosis control and mitigation strategy. The parasite Anisakis pegreffii (Nematoda: Anisakidae) realizes the final stage of its life-cycle within gastric chambers of aquatic mammals, causing mild-to-moderate granulomatous gastritis with eosinophilic infiltrate, to severe ulcerative gastritis with mixed inflammatory infiltrate, often associated with bacterial colonies. However, its interaction with the host microbiota remains unknown, and might reveal important aspects of parasite colonization and propagation within the final host.
Methods: MySeq Illumina sequencing was performed for the 16S rRNA gene from microbiota isolated from larvae, and uterus and gut of adult A. pegreffii parasitizing stranded striped dolphins (Stenella coeruleoalba). To assess the potential presence of Brucella ceti within isolated microbiota, Brucella-targeted real-time PCR was undertaken. In addition, TEM of the gastrointestinal tract of the infective third-stage (L3) and transitioning fourth-stage larvae (L4) was performed to characterize the morphological differences and the level of larval feeding activity.
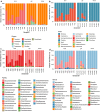
Results: In total, 230 distinct operational taxonomic units (OTUs) were identified across all samples (n = 20). The number of shared taxa was lower than the number of taxa found specifically in each parasite stage or organ. The dominant taxon was Mycoplasmataceae (genus Mycoplasma) in the gut and uterus of adult A. pegreffii, whereas Fusobacteriaceae (genus Cetobacterium) was the most abundant in 40% of larvae, alongside Mycoplasmataceae. No B. ceti DNA was detected in any of the microbiota isolates. TEM revealed differences in gut ultrastructure between L3 and L4, reflecting a feeble, most likely passive, level of feeding activity in L3.
Conclusions: Microbiota from L3 was more related to that of the gut rather than the uterus of adult A. pegreffii. Taxa of the larval microbiota showed qualitative and quantitative perturbations, likely reflecting the propagation through different environments during its life-cycle. This suggests an ontogenetic shift in the alpha and beta diversity of microbial communities from uterus-derived towards cetacean-derived microbiota. Although TEM did not reveal active L3 feeding, microbiota of the latter showed similarity to that of an actively feeding adult nematode.
Keywords: 16S rRNA gene sequencing; Anisakis pegreffii; Microbiota; Striped dolphin (Stenella coeruleoalba); TEM.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Marcogliese DJ. The role of zooplankton in the transmission of helminth parasites to fish. Rev Fish Biol Fish. 1995;5:336–371. doi: 10.1007/BF00043006. - DOI
-
- Anderson R. Host-parasite relations and evolution of the Metastrongyloidea (Nematoda) Mem Mus Natl Hist Nat Ser A Zool. 1982;123:129–132.
-
- Munn EA, Munn PD. Feeding and digestion. In: Lee DL, editor. The biology of nematodes. 1. London: Taylor & Francis; 2002. pp. 415–462.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

