Estrogen-independent molecular actions of mutant estrogen receptor 1 in endometrial cancer
- PMID: 31362937
- PMCID: PMC6724674
- DOI: 10.1101/gr.244780.118
Estrogen-independent molecular actions of mutant estrogen receptor 1 in endometrial cancer
Abstract
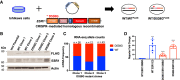
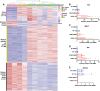
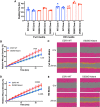
Estrogen receptor 1 (ESR1) mutations have been identified in hormone therapy-resistant breast cancer and primary endometrial cancer. Analyses in breast cancer suggest that mutant ESR1 exhibits estrogen-independent activity. In endometrial cancer, ESR1 mutations are associated with worse outcomes and less obesity, however, experimental investigation of these mutations has not been performed. Using a unique CRISPR/Cas9 strategy, we introduced the D538G mutation, a common endometrial cancer mutation that alters the ligand binding domain of ESR1, while epitope tagging the endogenous locus. We discovered estrogen-independent mutant ESR1 genomic binding that is significantly altered from wild-type ESR1. The D538G mutation impacted expression, including a large set of nonestrogen-regulated genes, and chromatin accessibility, with most affected loci bound by mutant ESR1. Mutant ESR1 is distinct from constitutive ESR1 activity because mutant-specific changes are not recapitulated with prolonged estrogen exposure. Overall, the D538G mutant ESR1 confers estrogen-independent activity while causing additional regulatory changes in endometrial cancer cells that are distinct from breast cancer cells.
© 2019 Blanchard et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
