Positive Selection and Functional Divergence at Meiosis Genes That Mediate Crossing Over Across the Drosophila Phylogeny
- PMID: 31362974
- PMCID: PMC6778797
- DOI: 10.1534/g3.119.400280
Positive Selection and Functional Divergence at Meiosis Genes That Mediate Crossing Over Across the Drosophila Phylogeny
Abstract
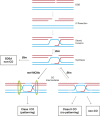
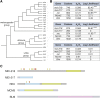
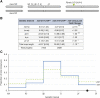
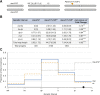
Meiotic crossing over ensures proper segregation of homologous chromosomes and generates genotypic diversity. Despite these functions, little is known about the genetic factors and population genetic forces involved in the evolution of recombination rate differences among species. The dicistronic meiosis gene, mei-217/mei-218, mediates most of the species differences in crossover rate and patterning during female meiosis between the closely related fruitfly species, Drosophila melanogaster and D. mauritiana The MEI-218 protein is one of several meiosis-specific mini-chromosome maintenance (mei-MCM) proteins that form a multi-protein complex essential to crossover formation, whereas the BLM helicase acts as an anti-crossover protein. Here we study the molecular evolution of five genes- mei-218, the other three known members of the mei-MCM complex, and Blm- over the phylogenies of three Drosophila species groups- melanogaster, obscura, and virilis We then use transgenic assays in D. melanogaster to test if molecular evolution at mei-218 has functional consequences for crossing over using alleles from the distantly related species D. pseudoobscura and D. virilis Our molecular evolutionary analyses reveal recurrent positive selection at two mei-MCM genes. Our transgenic assays show that sequence divergence among mei-218 alleles from D. melanogaster, D. pseudoobscura, and D. virilis has functional consequences for crossing over. In a D. melanogaster genetic background, the D. pseudoobscura mei-218 allele nearly rescues wildtype crossover rates but alters crossover patterning, whereas the D. virilis mei-218 allele conversely rescues wildtype crossover patterning but not crossover rates. These experiments demonstrate functional divergence at mei-218 and suggest that crossover rate and patterning are separable functions.
Keywords: Drosophila; crossing over; evolution; positive selection; recombination.
Copyright © 2019 Brand et al.
Figures
Similar articles
-
Molecular Evolution at a Meiosis Gene Mediates Species Differences in the Rate and Patterning of Recombination.Curr Biol. 2018 Apr 23;28(8):1289-1295.e4. doi: 10.1016/j.cub.2018.02.056. Epub 2018 Mar 29. Curr Biol. 2018. PMID: 29606420 Free PMC article.
-
Loss of Drosophila Mei-41/ATR Alters Meiotic Crossover Patterning.Genetics. 2018 Feb;208(2):579-588. doi: 10.1534/genetics.117.300634. Epub 2017 Dec 15. Genetics. 2018. PMID: 29247012 Free PMC article.
-
Meiotic MCM Proteins Promote and Inhibit Crossovers During Meiotic Recombination.Genetics. 2019 Jun;212(2):461-468. doi: 10.1534/genetics.119.302221. Epub 2019 Apr 26. Genetics. 2019. PMID: 31028111 Free PMC article.
-
[Genetic control of meiosis in Drozophila].Genetika. 2000 Oct;36(10):1301-21. Genetika. 2000. PMID: 11094742 Review. Russian.
-
The absence of crossovers on chromosome 4 in Drosophila melanogaster: Imperfection or interesting exception?Fly (Austin). 2017 Oct 2;11(4):253-259. doi: 10.1080/19336934.2017.1321181. Epub 2017 Apr 20. Fly (Austin). 2017. PMID: 28426351 Free PMC article. Review.
Cited by
-
Recombination landscape divergence between populations is marked by larger low-recombining regions in domesticated rye.Mol Biol Evol. 2022 Jun 11;39(6):msac131. doi: 10.1093/molbev/msac131. Online ahead of print. Mol Biol Evol. 2022. PMID: 35687854 Free PMC article.
-
Strong Signatures of Selection on Candidate Genes Underlying Core Speciation Mechanisms in Desert Tortoises.Mol Ecol Resour. 2025 Aug;25(6):e14098. doi: 10.1111/1755-0998.14098. Epub 2025 Mar 25. Mol Ecol Resour. 2025. PMID: 40129241 Free PMC article.
-
Diversification and recurrent adaptation of the synaptonemal complex in Drosophila.PLoS Genet. 2025 Jan 13;21(1):e1011549. doi: 10.1371/journal.pgen.1011549. eCollection 2025 Jan. PLoS Genet. 2025. PMID: 39804957 Free PMC article.
-
Taming the Turmoil Within: New Insights on the Containment of Transposable Elements.Trends Genet. 2020 Jul;36(7):474-489. doi: 10.1016/j.tig.2020.04.007. Epub 2020 May 27. Trends Genet. 2020. PMID: 32473745 Free PMC article. Review.
-
Genetics of recombination rate variation within and between species.J Evol Biol. 2025 Aug 2;38(7):851-860. doi: 10.1093/jeb/voae158. J Evol Biol. 2025. PMID: 39680417
References
-
- Baker W. K., 1958. Crossing over in heterochromatin. Am. Nat. 92: 59–60. 10.1086/282010 - DOI
-
- Baker B. S., and Hall J. C., 1976. Meiotic mutants: genetic control of meiotic recombination and chromosome segregation, pp. 352–434 in Genetics and Biology of Drosophila, edited by Novitski E., and Ashburner M. Academic Press, New York.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
