Common Variants in the Glycerol Kinase Gene Reduce Tuberculosis Drug Efficacy
- PMID: 31363023
- PMCID: PMC6667613
- DOI: 10.1128/mBio.00663-19
Common Variants in the Glycerol Kinase Gene Reduce Tuberculosis Drug Efficacy
Abstract
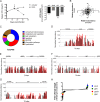
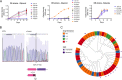
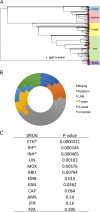
Despite the administration of multiple drugs that are highly effective in vitro, tuberculosis (TB) treatment requires prolonged drug administration and is confounded by the emergence of drug-resistant strains. To understand the mechanisms that limit antibiotic efficacy, we performed a comprehensive genetic study to identify Mycobacterium tuberculosis genes that alter the rate of bacterial clearance in drug-treated mice. Several functionally distinct bacterial genes were found to alter bacterial clearance, and prominent among these was the glpK gene that encodes the glycerol-3-kinase enzyme that is necessary for glycerol catabolism. Growth on glycerol generally increased the sensitivity of M. tuberculosis to antibiotics in vitro, and glpK-deficient bacteria persisted during antibiotic treatment in vivo, particularly during exposure to pyrazinamide-containing regimens. Frameshift mutations in a hypervariable homopolymeric region of the glpK gene were found to be a specific marker of multidrug resistance in clinical M. tuberculosis isolates, and these loss-of-function alleles were also enriched in extensively drug-resistant clones. These data indicate that frequently observed variation in the glpK coding sequence produces a drug-tolerant phenotype that can reduce antibiotic efficacy and may contribute to the evolution of resistance.IMPORTANCE TB control is limited in part by the length of antibiotic treatment needed to prevent recurrent disease. To probe mechanisms underlying survival under antibiotic pressure, we performed a genetic screen for M. tuberculosis mutants with altered susceptibility to treatment using the mouse model of TB. We identified multiple genes involved in a range of functions which alter sensitivity to antibiotics. In particular, we found glycerol catabolism mutants were less susceptible to treatment and that common variation in a homopolymeric region in the glpK gene was associated with drug resistance in clinical isolates. These studies indicate that reversible high-frequency variation in carbon metabolic pathways can produce phenotypically drug-tolerant clones and have a role in the development of resistance.
Keywords: Mycobacterium tuberculosis; antibiotic resistance; genetics.
Copyright © 2019 Bellerose et al.
Figures


Similar articles
-
Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance.Proc Natl Acad Sci U S A. 2019 Sep 24;116(39):19665-19674. doi: 10.1073/pnas.1907631116. Epub 2019 Sep 5. Proc Natl Acad Sci U S A. 2019. PMID: 31488707 Free PMC article.
-
Loss of glycerol catabolism confers carbon-source-dependent artemisinin resistance in Mycobacterium tuberculosis.Antimicrob Agents Chemother. 2024 Oct 8;68(10):e0064524. doi: 10.1128/aac.00645-24. Epub 2024 Aug 28. Antimicrob Agents Chemother. 2024. PMID: 39194262 Free PMC article.
-
[Frontier of mycobacterium research--host vs. mycobacterium].Kekkaku. 2005 Sep;80(9):613-29. Kekkaku. 2005. PMID: 16245793 Japanese.
-
Drug resistance, fitness and compensatory mutations in Mycobacterium tuberculosis.Tuberculosis (Edinb). 2021 Jul;129:102091. doi: 10.1016/j.tube.2021.102091. Epub 2021 May 21. Tuberculosis (Edinb). 2021. PMID: 34090078 Review.
-
Antibiotic resistance mechanisms in M. tuberculosis: an update.Arch Toxicol. 2016 Jul;90(7):1585-604. doi: 10.1007/s00204-016-1727-6. Epub 2016 May 9. Arch Toxicol. 2016. PMID: 27161440 Free PMC article. Review.
Cited by
-
Defining the mechanism of action of the nitrofuranyl piperazine HC2210 against Mycobacterium abscessus.NPJ Antimicrob Resist. 2025 Jun 14;3(1):55. doi: 10.1038/s44259-025-00124-0. NPJ Antimicrob Resist. 2025. PMID: 40517180 Free PMC article.
-
sRNA ncBCG427 activates the expression of target gene MSMEG_4757 to enhance the survival of Mycobacterium smegmatis through lipid metabolism in adverse environments.World J Microbiol Biotechnol. 2025 May 26;41(6):185. doi: 10.1007/s11274-025-04343-5. World J Microbiol Biotechnol. 2025. PMID: 40415042 Free PMC article.
-
Genome-wide screen identifies host loci that modulate Mycobacterium tuberculosis fitness in immunodivergent mice.G3 (Bethesda). 2023 Aug 30;13(9):jkad147. doi: 10.1093/g3journal/jkad147. G3 (Bethesda). 2023. PMID: 37405387 Free PMC article.
-
The rate and role of pseudogenes of the Mycobacterium tuberculosis complex.Microb Genom. 2022 Oct;8(10):mgen000876. doi: 10.1099/mgen.0.000876. Microb Genom. 2022. PMID: 36250787 Free PMC article.
-
Understanding Metabolic Regulation Between Host and Pathogens: New Opportunities for the Development of Improved Therapeutic Strategies Against Mycobacterium tuberculosis Infection.Front Cell Infect Microbiol. 2021 Mar 16;11:635335. doi: 10.3389/fcimb.2021.635335. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33796480 Free PMC article. Review.
References
-
- Fox W, Ellard GA, Mitchison DA. 1999. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council tuberculosis units, 1946-1986, with relevant subsequent publications. Int J Tuber Lung Dis 3:S231–S279. - PubMed
-
- World Health Organization. 2010. Treatment of tuberculosis guidelines, 4th ed World Health Organization, Geneva, Switzerland.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

