Chromosome Dynamics in Bacteria: Triggering Replication at the Opposite Location and Segregation in the Opposite Direction
- PMID: 31363028
- PMCID: PMC6667618
- DOI: 10.1128/mBio.01002-19
Chromosome Dynamics in Bacteria: Triggering Replication at the Opposite Location and Segregation in the Opposite Direction
Abstract
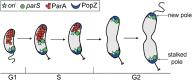
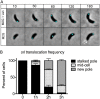
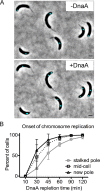
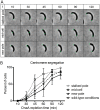
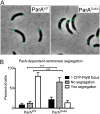
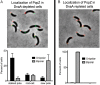
Maintaining the integrity of the genome is essential to cell survival. In the bacterium Caulobacter crescentus, the single circular chromosome exhibits a specific orientation in the cell, with the replication origin (ori) residing at the pole of the cell bearing a stalk. Upon initiation of replication, the duplicated centromere-like region parS and ori move rapidly to the opposite pole where parS is captured by a microdomain hosting a unique set of proteins that contribute to the identity of progeny cells. Many questions remain as to how this organization is maintained. In this study, we constructed strains of Caulobacter in which ori and the parS centromere can be induced to move to the opposite cell pole in the absence of chromosome replication, allowing us to ask whether once these chromosomal foci were positioned at the wrong pole, replication initiation and chromosome segregation can proceed in the opposite orientation. Our data reveal that DnaA can initiate replication and ParA can orchestrate segregation from either cell pole. The cell reconstructs the organization of its ParA gradient in the opposite orientation to segregate one replicated centromere from the new pole toward the stalked pole (i.e., opposite direction), while displaying no detectable viability defects. Thus, the unique polar microdomains exhibit remarkable flexibility in serving as a platform for directional chromosome segregation along the long axis of the cell.IMPORTANCE Bacteria can accomplish surprising levels of organization in the absence of membrane organelles by constructing subcellular asymmetric protein gradients. These gradients are composed of regulators that can either trigger or inhibit cell cycle events from distinct cell poles. In Caulobacter crescentus, the onset of chromosome replication and segregation from the stalked pole are regulated by asymmetric protein gradients. We show that the activators of chromosome replication and segregation are not restricted to the stalked pole and that their organization and directionality can be flipped in orientation. Our results also indicate that the subcellular location of key chromosomal loci play important roles in the establishment of the asymmetric organization of cell cycle regulators.
Keywords: Caulobacter crescentus; DnaA; ParA; centromere; chromosome replication; chromosome segregation.
Copyright © 2019 Meléndez et al.
Figures


References
-
- Hwang DS, Kornberg A. 1992. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem 267:23083–23086. - PubMed
-
- Sekimizu K, Bramhill D, Kornberg A. 1988. Sequential early stages in the in vitro initiation of replication at the origin of the Escherichia coli chromosome. J Biol Chem 263:7124–7130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

