Microbial Similarity between Students in a Common Dormitory Environment Reveals the Forensic Potential of Individual Microbial Signatures
- PMID: 31363029
- PMCID: PMC6667619
- DOI: 10.1128/mBio.01054-19
Microbial Similarity between Students in a Common Dormitory Environment Reveals the Forensic Potential of Individual Microbial Signatures
Abstract
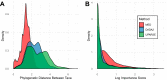
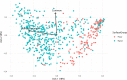
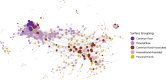
The microbiota of the built environment is an amalgamation of both human and environmental sources. While human sources have been examined within single-family households or in public environments, it is unclear what effect a large number of cohabitating people have on the microbial communities of their shared environment. We sampled the public and private spaces of a college dormitory, disentangling individual microbial signatures and their impact on the microbiota of common spaces. We compared multiple methods for marker gene sequence clustering and found that minimum entropy decomposition (MED) was best able to distinguish between the microbial signatures of different individuals and was able to uncover more discriminative taxa across all taxonomic groups. Further, weighted UniFrac- and random forest-based graph analyses uncovered two distinct spheres of hand- or shoe-associated samples. Using graph-based clustering, we identified spheres of interaction and found that connection between these clusters was enriched for hands, implicating them as a primary means of transmission. In contrast, shoe-associated samples were found to be freely interacting, with individual shoes more connected to each other than to the floors they interact with. Individual interactions were highly dynamic, with groups of samples originating from individuals clustering freely with samples from other individuals, while all floor and shoe samples consistently clustered together.IMPORTANCE Humans leave behind a microbial trail, regardless of intention. This may allow for the identification of individuals based on the "microbial signatures" they shed in built environments. In a shared living environment, these trails intersect, and through interaction with common surfaces may become homogenized, potentially confounding our ability to link individuals to their associated microbiota. We sought to understand the factors that influence the mixing of individual signatures and how best to process sequencing data to best tease apart these signatures.
Keywords: built environments; microbial ecology; microbial transmission.
Copyright © 2019 Richardson et al.
Figures


References
-
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vázquez-Baeza Y, Treuren WV, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, Gilbert JA. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–1052. doi:10.1126/science.1254529. - DOI - PMC - PubMed
-
- Meadow JF, Altrichter AE, Kembel SW, Kline J, Mhuireach G, Moriyama M, Northcutt D, O’Connor TK, Womack AM, Brown GZ, Green JL, Bohannan BJM. 2014. Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air 24:41–48. doi:10.1111/ina.12047. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
