Cellular Dynamics and Genomic Identity of Centromeres in Cereal Blast Fungus
- PMID: 31363034
- PMCID: PMC6667624
- DOI: 10.1128/mBio.01581-19
Cellular Dynamics and Genomic Identity of Centromeres in Cereal Blast Fungus
Abstract
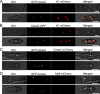
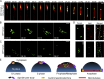
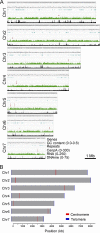
Precise kinetochore-microtubule interactions ensure faithful chromosome segregation in eukaryotes. Centromeres, identified as scaffolding sites for kinetochore assembly, are among the most rapidly evolving chromosomal loci in terms of the DNA sequence and length and organization of intrinsic elements. Neither the centromere structure nor the kinetochore dynamics is well studied in plant-pathogenic fungi. Here, we sought to understand the process of chromosome segregation in the rice blast fungus Magnaporthe oryzae High-resolution imaging of green fluorescent protein (GFP)-tagged inner kinetochore proteins CenpA and CenpC revealed unusual albeit transient declustering of centromeres just before anaphase separation of chromosomes in M. oryzae Strikingly, the declustered centromeres positioned randomly at the spindle midzone without an apparent metaphase plate per se Using CenpA chromatin immunoprecipitation followed by deep sequencing, all seven centromeres in M. oryzae were found to be regional, spanning 57-kb to 109-kb transcriptionally poor regions. Highly AT-rich and heavily methylated DNA sequences were the only common defining features of all the centromeres in rice blast. Lack of centromere-specific DNA sequence motifs or repetitive elements suggests an epigenetic specification of centromere function in M. oryzae PacBio genome assemblies and synteny analyses facilitated comparison of the centromeric/pericentromeric regions in distinct isolates of rice blast and wheat blast and in Magnaporthiopsis poae Overall, this study revealed unusual centromere dynamics and precisely identified the centromere loci in the top model fungal pathogens that belong to Magnaporthales and cause severe losses in the global production of food crops and turf grasses.IMPORTANCEMagnaporthe oryzae is an important fungal pathogen that causes a loss of 10% to 30% of the annual rice crop due to the devastating blast disease. In most organisms, kinetochores are clustered together or arranged at the metaphase plate to facilitate synchronized anaphase separation of sister chromatids in mitosis. In this study, we showed that the initially clustered kinetochores separate and position randomly prior to anaphase in M. oryzae Centromeres in M. oryzae occupy large genomic regions and form on AT-rich DNA without any common sequence motifs. Overall, this study identified atypical kinetochore dynamics and mapped functional centromeres in M. oryzae to define the roles of centromeric and pericentric boundaries in kinetochore assembly on epigenetically specified centromere loci. This study should pave the way for further understanding of the contribution of heterochromatin in genome stability and virulence of the blast fungus and its related species of high economic importance.
Keywords: CenpA; centromeres; chromosome structure; kinetochore dynamics; rice blast; wheat blast.
Copyright © 2019 Yadav et al.
Figures







Similar articles
-
Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells.J Cell Sci. 2003 Oct 1;116(Pt 19):4035-42. doi: 10.1242/jcs.00707. Epub 2003 Aug 19. J Cell Sci. 2003. PMID: 12928332
-
Gene Flow between Divergent Cereal- and Grass-Specific Lineages of the Rice Blast Fungus Magnaporthe oryzae.mBio. 2018 Feb 27;9(1):e01219-17. doi: 10.1128/mBio.01219-17. mBio. 2018. PMID: 29487238 Free PMC article.
-
Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis.Proc Natl Acad Sci U S A. 2008 Dec 16;105(50):19797-802. doi: 10.1073/pnas.0809770105. Epub 2008 Dec 5. Proc Natl Acad Sci U S A. 2008. PMID: 19060206 Free PMC article.
-
Artificial generation of centromeres and kinetochores to understand their structure and function.Exp Cell Res. 2020 Apr 15;389(2):111898. doi: 10.1016/j.yexcr.2020.111898. Epub 2020 Feb 6. Exp Cell Res. 2020. PMID: 32035949 Review.
-
Centromeric chromatin and its dynamics in plants.Plant J. 2015 Jul;83(1):4-17. doi: 10.1111/tpj.12875. Plant J. 2015. PMID: 25976696 Review.
Cited by
-
Functional and Comparative Analysis of Centromeres Reveals Clade-Specific Genome Rearrangements in Candida auris and a Chromosome Number Change in Related Species.mBio. 2021 May 11;12(3):e00905-21. doi: 10.1128/mBio.00905-21. mBio. 2021. PMID: 33975937 Free PMC article.
-
Genomic rearrangements generate hypervariable mini-chromosomes in host-specific isolates of the blast fungus.PLoS Genet. 2021 Feb 16;17(2):e1009386. doi: 10.1371/journal.pgen.1009386. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33591993 Free PMC article.
-
Pyricularia oryzae: Lab star and field scourge.Mol Plant Pathol. 2024 Apr;25(4):e13449. doi: 10.1111/mpp.13449. Mol Plant Pathol. 2024. PMID: 38619508 Free PMC article. Review.
-
Long transposon-rich centromeres in an oomycete reveal divergence of centromere features in Stramenopila-Alveolata-Rhizaria lineages.PLoS Genet. 2020 Mar 9;16(3):e1008646. doi: 10.1371/journal.pgen.1008646. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32150559 Free PMC article.
-
The Chromosome-Scale Genomes of Exserohilum rostratum and Bipolaris zeicola Pathogenic Fungi Causing Rice Spikelet Rot Disease.J Fungi (Basel). 2023 Jan 28;9(2):177. doi: 10.3390/jof9020177. J Fungi (Basel). 2023. PMID: 36836292 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

