Two PKA RIα holoenzyme states define ATP as an isoform-specific orthosteric inhibitor that competes with the allosteric activator, cAMP
- PMID: 31363049
- PMCID: PMC6697891
- DOI: 10.1073/pnas.1906036116
Two PKA RIα holoenzyme states define ATP as an isoform-specific orthosteric inhibitor that competes with the allosteric activator, cAMP
Abstract
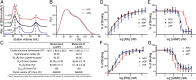
Protein kinase A (PKA) holoenzyme, comprised of a cAMP-binding regulatory (R)-subunit dimer and 2 catalytic (C)-subunits, is the master switch for cAMP-mediated signaling. Of the 4 R-subunits (RIα, RIβ, RIIα, RIIβ), RIα is most essential for regulating PKA activity in cells. Our 2 RIα2C2 holoenzyme states, which show different conformations with and without ATP, reveal how ATP/Mg2+ functions as a negative orthosteric modulator. Biochemical studies demonstrate how the removal of ATP primes the holoenzyme for cAMP-mediated activation. The opposing competition between ATP/cAMP is unique to RIα. In RIIβ, ATP serves as a substrate and facilitates cAMP-activation. The isoform-specific RI-holoenzyme dimer interface mediated by N3A-N3A' motifs defines multidomain cross-talk and an allosteric network that creates competing roles for ATP and cAMP. Comparisons to the RIIβ holoenzyme demonstrate isoform-specific holoenzyme interfaces and highlights distinct allosteric mechanisms for activation in addition to the structural diversity of the isoforms.
Keywords: allosteric and orthosteric regulation; cAMP; isoform-specific quaternary structure; protein kinase A; structural biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Knighton D. R., et al. , Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 (1991). - PubMed
-
- Johnson D. A., Akamine P., Radzio-Andzelm E., Madhusudan M., Taylor S. S., Dynamics of cAMP-dependent protein kinase. Chem. Rev. 101, 2243–2270 (2001). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

