UvrD helicase activation by MutL involves rotation of its 2B subdomain
- PMID: 31363055
- PMCID: PMC6697780
- DOI: 10.1073/pnas.1905513116
UvrD helicase activation by MutL involves rotation of its 2B subdomain
Abstract
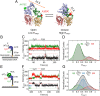
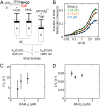
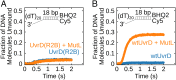
Escherichia coli UvrD is a superfamily 1 helicase/translocase that functions in DNA repair, replication, and recombination. Although a UvrD monomer can translocate along single-stranded DNA, self-assembly or interaction with an accessory protein is needed to activate its helicase activity in vitro. Our previous studies have shown that an Escherichia coli MutL dimer can activate the UvrD monomer helicase in vitro, but the mechanism is not known. The UvrD 2B subdomain is regulatory and can exist in extreme rotational conformational states. By using single-molecule FRET approaches, we show that the 2B subdomain of a UvrD monomer bound to DNA exists in equilibrium between open and closed states, but predominantly in an open conformation. However, upon MutL binding to a UvrD monomer-DNA complex, a rotational conformational state is favored that is intermediate between the open and closed states. Parallel kinetic studies of MutL activation of the UvrD helicase and of MutL-dependent changes in the UvrD 2B subdomain show that the transition from an open to an intermediate 2B subdomain state is on the pathway to helicase activation. We further show that MutL is unable to activate the helicase activity of a chimeric UvrD containing the 2B subdomain of the structurally similar Rep helicase. Hence, MutL activation of the monomeric UvrD helicase is regulated specifically by its 2B subdomain.
Keywords: activation; conformational selection; helicase; mismatch repair; single molecule fluorescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Iyer R. R., Pluciennik A., Burdett V., Modrich P. L., DNA mismatch repair: Functions and mechanisms. Chem. Rev. 106, 302–323 (2006). - PubMed
-
- Sancar A., DNA excision repair. Annu. Rev. Biochem. 65, 43–81 (1996). - PubMed
-
- Heller R. C., Marians K. J., Non-replicative helicases at the replication fork. DNA Repair (Amst.) 6, 945–952 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

