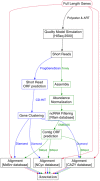
To assemble or not to resemble-A validated Comparative Metatranscriptomics Workflow (CoMW)
- PMID: 31363751
- PMCID: PMC6667343
- DOI: 10.1093/gigascience/giz096
To assemble or not to resemble-A validated Comparative Metatranscriptomics Workflow (CoMW)
Abstract
Background: Metatranscriptomics has been used widely for investigation and quantification of microbial communities' activity in response to external stimuli. By assessing the genes expressed, metatranscriptomics provides an understanding of the interactions between different major functional guilds and the environment. Here, we present a de novo assembly-based Comparative Metatranscriptomics Workflow (CoMW) implemented in a modular, reproducible structure. Metatranscriptomics typically uses short sequence reads, which can either be directly aligned to external reference databases ("assembly-free approach") or first assembled into contigs before alignment ("assembly-based approach"). We also compare CoMW (assembly-based implementation) with an assembly-free alternative workflow, using simulated and real-world metatranscriptomes from Arctic and temperate terrestrial environments. We evaluate their accuracy in precision and recall using generic and specialized hierarchical protein databases.
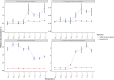
Results: CoMW provided significantly fewer false-positive results, resulting in more precise identification and quantification of functional genes in metatranscriptomes. Using the comprehensive database M5nr, the assembly-based approach identified genes with only 0.6% false-positive results at thresholds ranging from inclusive to stringent compared with the assembly-free approach, which yielded up to 15% false-positive results. Using specialized databases (carbohydrate-active enzyme and nitrogen cycle), the assembly-based approach identified and quantified genes with 3-5 times fewer false-positive results. We also evaluated the impact of both approaches on real-world datasets.
Conclusions: We present an open source de novo assembly-based CoMW. Our benchmarking findings support assembling short reads into contigs before alignment to a reference database because this provides higher precision and minimizes false-positive results.
Keywords: alignment; assembly; benchmarking; false-positive results; metatranscriptomics; precision; recall.
© The Author(s) 2019. Published by Oxford University Press.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

