Amplicon-based microbiome study highlights the loss of diversity and the establishment of a set of species in patients with dentin caries
- PMID: 31365560
- PMCID: PMC6668773
- DOI: 10.1371/journal.pone.0219714
Amplicon-based microbiome study highlights the loss of diversity and the establishment of a set of species in patients with dentin caries
Abstract
Objectives: To elicit patterns in pathogenic biofilm composition we characterized the oral microbiome present in patients with dentin caries in comparison to healthy subjects.
Methods: 16S amplicon sequencing was used to analyse a total of 56 patients; 19 samples of carious dentin (pooled from at least three teeth) and 37 supragingival samples (pooled from three healthy tooth surfaces). Oral and periodontal status and socio-demographic parameters were recorded. Group assignment, smoking and further socio-demographic parameters were used as explanatory variables in the microbiome composition analysis.
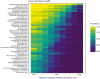
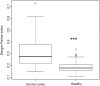
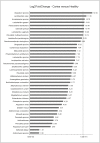
Results: Overall, a total of 4,110,020 DNA high-quality sequences were yielded. Using a threshold of similarity >97% for assigning operational taxonomic units (OTU), a total of 1,537 OTUs were identified. PERMANOVA showed significant differences in microbiome composition between the groups caries/healthy (p = 0.001), smoking/non-smoking (p = 0.007) and fluoride intake during childhood yes/no (tablets p = 0.003, salt p = 0.023). The healthy microbiome had a significantly higher diversity (alpha diversity, p<0.001) and a lower dominance (Berger-Parker index, p<0.001). It was dominated by Fusobacteria. A linear discriminant analysis effect size (LEfSe) yielded a set of 39 OTUs being more abundant in carious dentin samples, including Atopobium spp. (14.9 log2FoldChange), Lactobacillus casei (11.6), Acinetobacter spp. (10.8), Lactobacillus gasseri (10.6), Parascardovia denticolens (10.5), Olsenella profusa (10.4), and others. Also Propionibacterium acidifaciens (7.2) and Streptococcus mutans (5.2) were overabundant in caries lesions.
Conclusions: The healthy microbiome was highly diverse. The advanced caries microbiome was dominated by a set of carious associated bacteria where S. mutans played only a minor role. Smoking and fluoride intake during childhood influenced the microbiome composition significantly.
Clinical significance: The presented investigation adds knowledge to the still not fully comprehended patterns of oral microbiomes in caries compared with oral health. By analysing the genetics of biofilm samples from oral health and severe tooth decay we found distinct discriminating species which could be targets for future therapeutic approaches.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Hurley E, Barrett MPJ, Kinirons M, Whelton H, Ryan CA, Stanton C, et al. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health. 2019. January;19(1):13 10.1186/s12903-018-0693-1 . - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

