A urinary Common Rejection Module (uCRM) score for non-invasive kidney transplant monitoring
- PMID: 31365568
- PMCID: PMC6668802
- DOI: 10.1371/journal.pone.0220052
A urinary Common Rejection Module (uCRM) score for non-invasive kidney transplant monitoring
Abstract
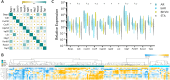
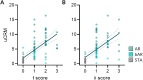
A Common Rejection Module (CRM) consisting of 11 genes expressed in allograft biopsies was previously reported to serve as a biomarker for acute rejection (AR), correlate with the extent of graft injury, and predict future allograft damage. We investigated the use of this gene panel on the urine cell pellet of kidney transplant patients. Urinary cell sediments collected from patients with biopsy-confirmed acute rejection, borderline AR (bAR), BK virus nephropathy (BKVN), and stable kidney grafts with normal protocol biopsies (STA) were analyzed for expression of these 11 genes using quantitative polymerase chain reaction (qPCR). We assessed these 11 CRM genes for their abundance, autocorrelation, and individual expression levels. Expression of 10/11 genes were elevated in AR when compared to STA. Psmb9 and Cxcl10could classify AR versus STA as accurately as the 11-gene model (sensitivity = 93.6%, specificity = 97.6%). A uCRM score, based on the geometric mean of the expression levels, could distinguish AR from STA with high accuracy (AUC = 0.9886) and correlated specifically with histologic measures of tubulitis and interstitial inflammation rather than tubular atrophy, glomerulosclerosis, intimal proliferation, tubular vacuolization or acute glomerulitis. This urine gene expression-based score may enable the non-invasive and quantitative monitoring of AR.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: J.Y.C.Y. and M.M.S. are co-founders of KIT Bio, Inc. The research conducted in this manuscript is unrelated to the commercial activities of this company. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures



References
-
- Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. The New England journal of medicine. 1999;341(23):1725–30. 10.1056/NEJM199912023412303 . - DOI - PubMed
-
- Laupacis A, Keown P, Pus N, Krueger H, Ferguson B, Wong C, et al. A study of the quality of life and cost-utility of renal transplantation. Kidney international. 1996;50(1):235–42. . - PubMed
-
- Lodhi SA, Lamb KE, Meier-Kriesche HU. Improving long-term outcomes for transplant patients: making the case for long-term disease-specific and multidisciplinary research. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11(10):2264–5. 10.1111/j.1600-6143.2011.03713.x . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

