Differential Inhibition of Target Gene Expression by Human microRNAs
- PMID: 31366019
- PMCID: PMC6721455
- DOI: 10.3390/cells8080791
Differential Inhibition of Target Gene Expression by Human microRNAs
Abstract
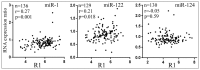
microRNAs (miRNAs) exert their functions by repressing the expression of their target genes, but most miRNA target genes are unknown, and the degree to which a miRNA differentially inhibits the expression of its targets is underappreciated. We selected human miR-1, miR-122, and miR-124 as representatives to investigate the reliability of miRNA target predictions and examine how miRNAs suppress their targets. We constructed miRNA target gene reporter libraries based on prediction programs TargetScan, miRanda, and PicTar, and performed large-scale reporter assays to directly evaluate whether and how strongly a predicted target gene is repressed by its miRNA. We then performed statistical analyses to examine parameters that contributed to the miRNA inhibition of target genes. We found that the three programs have approximately 72-85% success rates in predicting genuine targets and that the miRNA inhibition of different targets varies in extent. We also identified parameters that could predict the degrees of miRNA repression, and further showed that differential miR-124 repression might contribute to differential gene expression in vivo. Our studies systematically investigated hundreds of miRNA target genes, shed light on factors influencing miRNA functions, and suggested a new mechanism by which differential target repression by miRNAs regulates endogenous gene expression.
Keywords: differential gene expression; differential gene suppression; miRNA; miRNA target gene; target gene prediction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

