Application of a Substrate-Mediated Selection with c-Src Tyrosine Kinase to a DNA-Encoded Chemical Library
- PMID: 31366048
- PMCID: PMC6695731
- DOI: 10.3390/molecules24152764
Application of a Substrate-Mediated Selection with c-Src Tyrosine Kinase to a DNA-Encoded Chemical Library
Abstract
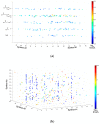
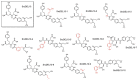
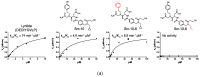
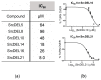
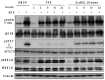
As aberrant activity of protein kinases is observed in many disease states, these enzymes are common targets for therapeutics and detection of activity levels. The development of non-natural protein kinase substrates offers an approach to protein substrate competitive inhibitors, a class of kinase inhibitors with promise for improved specificity. Also, kinase activity detection approaches would benefit from substrates with improved activity and specificity. Here, we apply a substrate-mediated selection to a peptidomimetic DNA-encoded chemical library for enrichment of molecules that can be phosphorylated by the protein tyrosine kinase, c-Src. Several substrates were identified and characterized for activity. A lead compound (SrcDEL10) showed both the ability to serve as a substrate and to promote ATP hydrolysis by the kinase. In inhibition assays, compounds displayed IC50's ranging from of 8-100 µM. NMR analysis of SrcDEL10 bound to the c-Src:ATP complex was conducted to characterize the binding mode. An ester derivative of the lead compound demonstrated cellular activity with inhibition of Src-dependent signaling in cell culture. Together, the results show the potential for substrate-mediated selections of DNA-encoded libraries to discover molecules with functions other than simple protein binding and offer a new discovery method for development of synthetic tyrosine kinase substrates.
Keywords: DNA-encoded chemical library; c-Src; protein tyrosine kinases; substrate-mediated selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

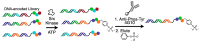



Similar articles
-
Substrate activity screening with kinases: discovery of small-molecule substrate-competitive c-Src inhibitors.Angew Chem Int Ed Engl. 2014 Jul 1;53(27):7010-3. doi: 10.1002/anie.201311096. Epub 2014 May 2. Angew Chem Int Ed Engl. 2014. PMID: 24797781 Free PMC article.
-
Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library.Chembiochem. 2017 May 4;18(9):864-871. doi: 10.1002/cbic.201600573. Epub 2017 Feb 8. Chembiochem. 2017. PMID: 28056160
-
Structural basis of Src tyrosine kinase inhibition with a new class of potent and selective trisubstituted purine-based compounds.Chem Biol Drug Des. 2006 Jan;67(1):46-57. doi: 10.1111/j.1747-0285.2005.00316.x. Chem Biol Drug Des. 2006. PMID: 16492148
-
Isolation of novel Src substrates.Biochem Soc Trans. 2003 Feb;31(Pt 1):25-8. doi: 10.1042/bst0310025. Biochem Soc Trans. 2003. PMID: 12546647 Review.
-
Regulation, substrates and functions of src.Biochim Biophys Acta. 1996 Jun 7;1287(2-3):121-49. doi: 10.1016/0304-419x(96)00003-0. Biochim Biophys Acta. 1996. PMID: 8672527 Review.
Cited by
-
DNA-encoded libraries (DELs): a review of on-DNA chemistries and their output.RSC Adv. 2021 Jan 19;11(4):2359-2376. doi: 10.1039/d0ra09889b. eCollection 2021 Jan 6. RSC Adv. 2021. PMID: 35424149 Free PMC article. Review.
-
Selection methods for proximity-dependent enrichment of ligands from DNA-encoded libraries using enzymatic fusion proteins.Chem Sci. 2022 Nov 15;14(2):245-250. doi: 10.1039/d2sc05495g. eCollection 2023 Jan 4. Chem Sci. 2022. PMID: 36687357 Free PMC article.
-
The protein kinase CK1: Inhibition, activation, and possible allosteric modulation.Front Mol Biosci. 2022 Aug 24;9:916232. doi: 10.3389/fmolb.2022.916232. eCollection 2022. Front Mol Biosci. 2022. PMID: 36090057 Free PMC article. Review.
-
Small-molecule discovery through DNA-encoded libraries.Nat Rev Drug Discov. 2023 Sep;22(9):699-722. doi: 10.1038/s41573-023-00713-6. Epub 2023 Jun 16. Nat Rev Drug Discov. 2023. PMID: 37328653 Free PMC article. Review.
-
DNA-encoded probe-based assay for profiling plant kinase activities.PNAS Nexus. 2024 Jul 19;3(7):pgae281. doi: 10.1093/pnasnexus/pgae281. eCollection 2024 Jul. PNAS Nexus. 2024. PMID: 39045014 Free PMC article.
References
-
- Cuozzo J.W., Centrella P.A., Gikunju D., Habeshian S., Hupp C.D., Keefe A.D., Sigel E.A., Soutter H.H., Thomson H.A., Zhang Y., et al. Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library. ChemBioChem. 2017;18:864–871. doi: 10.1002/cbic.201600573. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

