BST2/Tetherin Overexpression Modulates Morbillivirus Glycoprotein Production to Inhibit Cell-Cell Fusion
- PMID: 31366072
- PMCID: PMC6723339
- DOI: 10.3390/v11080692
BST2/Tetherin Overexpression Modulates Morbillivirus Glycoprotein Production to Inhibit Cell-Cell Fusion
Abstract
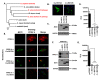
The measles virus (MeV), a member of the genus Morbillivirus, is an established pathogen of humans. A key feature of morbilliviruses is their ability to spread by virus-cell and cell-cell fusion. The latter process, which leads to syncytia formation in vitro and in vivo, is driven by the viral fusion (F) and haemagglutinin (H) glycoproteins. In this study, we demonstrate that MeV glycoproteins are sensitive to inhibition by bone marrow stromal antigen 2 (BST2/Tetherin/CD317) proteins. BST2 overexpression causes a large reduction in MeV syncytia expansion. Using quantitative cell-cell fusion assays, immunolabeling, and biochemistry we further demonstrate that ectopically expressed BST2 directly inhibits MeV cell-cell fusion. This restriction is mediated by the targeting of the MeV H glycoprotein, but not other MeV proteins. Using truncation mutants, we further establish that the C-terminal glycosyl-phosphatidylinositol (GPI) anchor of BST2 is required for the restriction of MeV replication in vitro and cell-cell fusion. By extending our study to the ruminant morbillivirus peste des petits ruminants virus (PPRV) and its natural host, sheep, we also confirm this is a broad and cross-species specific phenotype.
Keywords: BST2; MeV; Morbillivirus; PPRV; haemagluttinin; measles; tetherin.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures
Similar articles
-
Structure-Guided Identification of a Nonhuman Morbillivirus with Zoonotic Potential.J Virol. 2018 Nov 12;92(23):e01248-18. doi: 10.1128/JVI.01248-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30232185 Free PMC article.
-
Productive replication of peste des petits ruminants virus Nigeria 75/1 vaccine strain in vero cells correlates with inefficiency of maturation of the viral fusion protein.Virus Res. 2019 Aug;269:197634. doi: 10.1016/j.virusres.2019.05.012. Epub 2019 May 23. Virus Res. 2019. PMID: 31129173
-
Immune responses in goats to recombinant hemagglutinin-neuraminidase glycoprotein of Peste des petits ruminants virus: identification of a T cell determinant.Vaccine. 2001 Sep 14;19(32):4816-23. doi: 10.1016/s0264-410x(01)00210-9. Vaccine. 2001. PMID: 11535334
-
Host Cellular Receptors for the Peste des Petits Ruminant Virus.Viruses. 2019 Aug 8;11(8):729. doi: 10.3390/v11080729. Viruses. 2019. PMID: 31398809 Free PMC article. Review.
-
Morbillivirus vaccines: recent successes and future hopes.Vaccine. 2014 May 30;32(26):3155-61. doi: 10.1016/j.vaccine.2014.03.053. Epub 2014 Apr 3. Vaccine. 2014. PMID: 24703852 Free PMC article. Review.
Cited by
-
Virus-Mediated Cell-Cell Fusion.Int J Mol Sci. 2020 Dec 17;21(24):9644. doi: 10.3390/ijms21249644. Int J Mol Sci. 2020. PMID: 33348900 Free PMC article. Review.
-
Measles Encephalitis: Towards New Therapeutics.Viruses. 2019 Nov 2;11(11):1017. doi: 10.3390/v11111017. Viruses. 2019. PMID: 31684034 Free PMC article. Review.
-
Murine BST2/tetherin promotes measles virus infection of neurons.Virology. 2021 Nov;563:38-43. doi: 10.1016/j.virol.2021.08.005. Epub 2021 Aug 15. Virology. 2021. PMID: 34416448 Free PMC article.
-
Micro-fusion inhibition tests: quantifying antibody neutralization of virus-mediated cell-cell fusion.J Gen Virol. 2021 Jan;102(1):jgv001506. doi: 10.1099/jgv.0.001506. Epub 2020 Oct 15. J Gen Virol. 2021. PMID: 33054904 Free PMC article.
-
Retroviral Restriction Factors and Their Viral Targets: Restriction Strategies and Evolutionary Adaptations.Microorganisms. 2020 Dec 11;8(12):1965. doi: 10.3390/microorganisms8121965. Microorganisms. 2020. PMID: 33322320 Free PMC article. Review.
References
-
- World Health Orgnization Measles (Fact Sheet No 286) [(accessed on 24 July 2019)]; Available online: www.who.int/mediacentre/factsheets/fs286/en/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

