Systematic Analysis of MYB Family Genes in Potato and Their Multiple Roles in Development and Stress Responses
- PMID: 31366107
- PMCID: PMC6723670
- DOI: 10.3390/biom9080317
Systematic Analysis of MYB Family Genes in Potato and Their Multiple Roles in Development and Stress Responses
Abstract
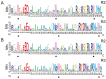
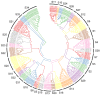
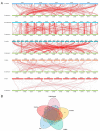
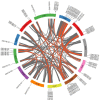
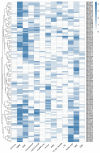
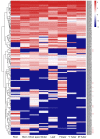
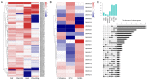
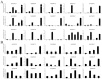
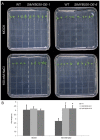
The MYB proteins represent a large family of transcription factors and play important roles in development, senescence, and stress responses in plants. In the current study, 233 MYB transcription factor-encoding genes were identified and analyzed in the potato genome, including 119 R1-MYB, 112 R2R3-MYB, and two R1R2R3-MYB members. R2R3-MYB is the most abundant MYB subclass and potato R2R3-MYB members together with their Arabidopsis homologs were divided into 35 well-supported subgroups as the result of phylogenetic analyses. Analyses on gene structure and protein motif revealed that members from the same subgroup shared similar exon/intron and motif organization, further supporting the results of phylogenetic analyses. Evolution of the potato MYB family was studied via syntenic analysis. Forty-one pairs of StMYB genes were predicted to have arisen from tandem or segmental duplication events, which played important roles in the expansion of the StMYB family. Expression profiling revealed that the StMYB genes were expressed in various tissues and several StMYB genes were identified to be induced by different stress conditions. Notably, StMYB030 was found to act as the homolog of AtMYB44 and was significantly up-regulated by salt and drought stress treatments. Furthermore, overexpression of StMYB030 in Arabidopsis enhanced salt stress tolerance of transgenic plants. The results from this study provided information for further functional analysis and for crop improvements through genetic manipulation of these StMYB genes.
Keywords: MYB; abiotic stress; biotic stress; leaf senescence; potato.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ogata K., Kanei-Ishii C., Sasaki M., Hatanaka H., Nagadoi A., Enari M., Nakamura H., Nishimura Y., Ishii S., Sarai A. The cavity in the hydrophobic core of MYB DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Mol. Biol. 1996;3:178–187. doi: 10.1038/nsb0296-178. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

