Therapeutically relevant engraftment of a CRISPR-Cas9-edited HSC-enriched population with HbF reactivation in nonhuman primates
- PMID: 31366580
- PMCID: PMC8407476
- DOI: 10.1126/scitranslmed.aaw3768
Therapeutically relevant engraftment of a CRISPR-Cas9-edited HSC-enriched population with HbF reactivation in nonhuman primates
Abstract
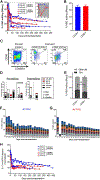
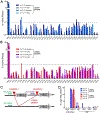
Reactivation of fetal hemoglobin (HbF) is being pursued as a treatment strategy for hemoglobinopathies. Here, we evaluated the therapeutic potential of hematopoietic stem and progenitor cells (HSPCs) edited with the CRISPR-Cas9 nuclease platform to recapitulate naturally occurring mutations identified in individuals who express increased amounts of HbF, a condition known as hereditary persistence of HbF. CRISPR-Cas9 treatment and transplantation of HSPCs purified on the basis of surface expression of the CD34 receptor in a nonhuman primate (NHP) autologous transplantation model resulted in up to 30% engraftment of gene-edited cells for >1 year. Edited cells effectively and stably reactivated HbF, as evidenced by up to 18% HbF-expressing erythrocytes in peripheral blood. Similar results were obtained by editing highly enriched stem cells, defined by the markers CD34+CD90+CD45RA-, allowing for a 10-fold reduction in the number of transplanted target cells, thus considerably reducing the need for editing reagents. The frequency of engrafted, gene-edited cells persisting in vivo using this approach may be sufficient to ameliorate the phenotype for a number of genetic diseases.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

