Genomic landscape of neutrophilic leukemias of ambiguous diagnosis
- PMID: 31366621
- PMCID: PMC6742922
- DOI: 10.1182/blood.2019000611
Genomic landscape of neutrophilic leukemias of ambiguous diagnosis
Abstract
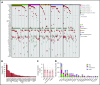
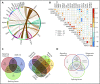
Chronic neutrophilic leukemia (CNL), atypical chronic myeloid leukemia (aCML), and myelodysplastic/myeloproliferative neoplasms, unclassifiable (MDS/MPN-U) are a group of rare and heterogeneous myeloid disorders. There is strong morphologic resemblance among these distinct diagnostic entities as well as a lack of specific molecular markers and limited understanding of disease pathogenesis, which has made diagnosis challenging in certain cases. The treatment has remained empirical, resulting in dismal outcomes. We, therefore, performed whole-exome and RNA sequencing of these rare hematologic malignancies and present the most complete survey of the genomic landscape of these diseases to date. We observed a diversity of combinatorial mutational patterns that generally do not cluster within any one diagnosis. Gene expression analysis reveals enrichment, but not cosegregation, of clinical and genetic disease features with transcriptional clusters. In conclusion, these groups of diseases represent a continuum of related diseases rather than discrete diagnostic entities.
© 2019 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: J.W.T. has received research support from Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Seattle Genetics, Syros, and Takeda and is co-founder of Vivid Biosciences. B.J.D. is on the scientific advisory board of Aileron Therapeutics, ALLCRON, Cepheid, Gilead Sciences, Vivid Biosciences, Celgene, and Baxalta (inactive); is on the scientific advisory board of and owns stock in Aptose Biosciences, Blueprint Medicines, Beta Cat, GRAIL, Third Coast Therapeutics, and CTI BioPharma (inactive); is a scientific founder of and owns stock in MolecularMD; is on the board of directors and owns stock in Amgen; is on the board of directors of Burroughs Wellcome Fund and CureOne; is on the Joint Steering Committee of Beat AML LLS; and receives clinical trial funding from Novartis, Bristol-Myers Squibb, and Pfizer and royalties from patent 6958335 (Novartis exclusive license), Oregon Health & Science University, and Dana-Farber Cancer Institute (1 Merck exclusive license). K.-H.T.D. is a member of the advisory board of Incyte and received funding from Incyte for research study. The remaining authors declare no competing financial interests.
Figures






Comment in
-
Illuminating neutrophilic myeloid neoplasms.Blood. 2019 Sep 12;134(11):846-848. doi: 10.1182/blood.2019002302. Blood. 2019. PMID: 31515226 No abstract available.
References
-
- Bupathi M, Tiu RV, Maciejewski JP. Myelodysplastic/Myeloproliferative Neoplasms, 2nd ed Myelodysplastic Syndr; 2013:111-126
-
- Deininger MWN, Tyner JW, Solary E. Turning the tide in myelodysplastic/myeloproliferative neoplasms. Nat Rev Cancer. 2017;17(7):425-440. - PubMed
-
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

