The cancer-associated, gain-of-function TP53 variant P152Lp53 activates multiple signaling pathways implicated in tumorigenesis
- PMID: 31366730
- PMCID: PMC6755804
- DOI: 10.1074/jbc.RA118.007265
The cancer-associated, gain-of-function TP53 variant P152Lp53 activates multiple signaling pathways implicated in tumorigenesis
Abstract
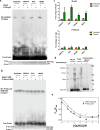
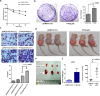
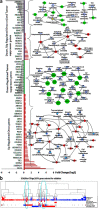
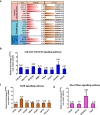
TP53 is the most frequently mutated tumor suppressor gene in many cancers, yet biochemical characterization of several of its reported mutations with probable biological significance have not been accomplished enough. Specifically, missense mutations in TP53 can contribute to tumorigenesis through gain-of-function of biochemical and biological properties that stimulate tumor growth. Here, we identified a relatively rare mutation leading to a proline to leucine substitution (P152L) in TP53 at the very end of its DNA-binding domain (DBD) in a sample from an Indian oral cancer patient. Although the P152Lp53 DBD alone bound to DNA, the full-length protein completely lacked binding ability at its cognate DNA motifs. Interestingly, P152Lp53 could efficiently tetramerize, and the mutation had only a limited impact on the structure and stability of full-length p53. Significantly, when we expressed this variant in a TP53-null cell line, it induced cell motility, proliferation, and invasion compared with a vector-only control. Also, enhanced tumorigenic potential was observed when P152Lp53-expressing cells were xenografted into nude mice. Investigating the effects of P152Lp53 expression on cellular pathways, we found that it is associated with up-regulation of several pathways, including cell-cell and cell-extracellular matrix signaling, epidermal growth factor receptor signaling, and Rho-GTPase signaling, commonly active in tumorigenesis and metastasis. Taken together, our findings provide a detailed account of the biochemical and cellular alterations associated with the cancer-associated P152Lp53 variant and establish it as a gain-of-function TP53 variant.
Keywords: DNA-binding protein; P152Lp53; carcinogenesis; gain-of-function mutant; gene expression; p53 tetramerization; transcriptional regulator; tumor cell biology; tumor suppressor gene.
© 2019 Singh et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
Gain-of-function p53 activates multiple signaling pathways to induce oncogenicity in lung cancer cells.Mol Oncol. 2017 Jun;11(6):696-711. doi: 10.1002/1878-0261.12068. Epub 2017 May 8. Mol Oncol. 2017. PMID: 28423230 Free PMC article.
-
The activating transcription factor 3 protein suppresses the oncogenic function of mutant p53 proteins.J Biol Chem. 2014 Mar 28;289(13):8947-59. doi: 10.1074/jbc.M113.503755. Epub 2014 Feb 19. J Biol Chem. 2014. PMID: 24554706 Free PMC article.
-
p53‑p72‑Δ225‑331‑V31I identified in a cholangiocarcinoma cell line promotes migration and invasiveness via the downregulation of claudin‑1 expression and the activation of Cdc42.Oncol Rep. 2021 Jan;45(1):368-378. doi: 10.3892/or.2020.7827. Epub 2020 Oct 27. Oncol Rep. 2021. PMID: 33416138
-
Mutant p53 on the Path to Metastasis.Trends Cancer. 2020 Jan;6(1):62-73. doi: 10.1016/j.trecan.2019.11.004. Epub 2019 Dec 16. Trends Cancer. 2020. PMID: 31952783 Free PMC article. Review.
-
Lung-Enriched Mutations in the p53 Tumor Suppressor: A Paradigm for Tissue-Specific Gain of Oncogenic Function.Mol Cancer Res. 2019 Jan;17(1):3-9. doi: 10.1158/1541-7786.MCR-18-0357. Epub 2018 Sep 17. Mol Cancer Res. 2019. PMID: 30224539 Free PMC article. Review.
Cited by
-
The genomic architectures of tumour-adjacent tissues, plasma and saliva reveal evolutionary underpinnings of relapse in head and neck squamous cell carcinoma.Br J Cancer. 2021 Sep;125(6):854-864. doi: 10.1038/s41416-021-01464-0. Epub 2021 Jul 6. Br J Cancer. 2021. PMID: 34230611 Free PMC article.
-
Impact of p53-associated acute myeloid leukemia hallmarks on metabolism and the immune environment.Front Pharmacol. 2024 Aug 5;15:1409210. doi: 10.3389/fphar.2024.1409210. eCollection 2024. Front Pharmacol. 2024. PMID: 39161899 Free PMC article. Review.
-
KRAS Mutation Subtypes and Their Association with Other Driver Mutations in Oncogenic Pathways.Cells. 2024 Jul 19;13(14):1221. doi: 10.3390/cells13141221. Cells. 2024. PMID: 39056802 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

