Cholera toxin promotes pathogen acquisition of host-derived nutrients
- PMID: 31367037
- PMCID: PMC6727848
- DOI: 10.1038/s41586-019-1453-3
Cholera toxin promotes pathogen acquisition of host-derived nutrients
Abstract
Vibrio cholerae is the causative agent of cholera, a potentially lethal enteric bacterial infection1. Cholera toxin (CTX), a protein complex that is secreted by V. cholerae, is required for V. cholerae to cause severe disease. CTX is also thought to promote transmission of the organism, as infected individuals shed many litres of diarrhoeal fluid that typically contains in excess of 1011 organisms per litre. How the pathogen is able to reach such high concentrations in the intestine during infection remains poorly understood. Here we show that CTX increases pathogen growth and induces a distinct V. cholerae transcriptomic signature that is indicative of an iron-depleted gut niche. During infection, bacterial pathogens need to acquire iron, which is an essential nutrient for growth2. Most iron in the mammalian host is found in a chelated form within the porphyrin structure of haem, and the ability to use haem as a source of iron is genetically encoded by V. cholerae3. We show that the genes that enable V. cholerae to obtain iron via haem and vibriobactin confer a growth advantage to the pathogen only when CTX is produced. Furthermore, we found that CTX-induced congestion of capillaries in the terminal ileum correlated with an increased bioavailability of luminal haem. CTX-induced disease in the ileum also led to increased concentrations of long-chain fatty acids and L-lactate metabolites in the lumen, as well as the upregulation of V. cholerae genes that encode enzymes of the tricarboxylic acid (TCA) cycle that contain iron-sulfur clusters. Genetic analysis of V. cholerae suggested that pathogen growth was dependent on the uptake of haem and long-chain fatty acids during infection, but only in a strain capable of producing CTX in vivo. We conclude that CTX-induced disease creates an iron-depleted metabolic niche in the gut, which selectively promotes the growth of V. cholerae through the acquisition of host-derived haem and fatty acids.
Figures

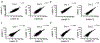
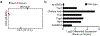






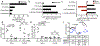
References
REFERENCES (Online-only)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

