Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis
- PMID: 31367040
- PMCID: PMC6707815
- DOI: 10.1038/s41586-019-1450-6
Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis
Abstract
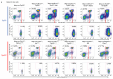
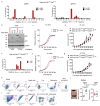
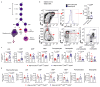
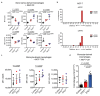
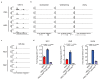
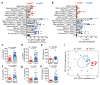
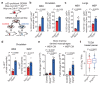
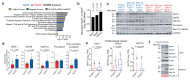
Cancer-associated systemic inflammation is strongly linked to poor disease outcome in patients with cancer1,2. For most human epithelial tumour types, high systemic neutrophil-to-lymphocyte ratios are associated with poor overall survival3, and experimental studies have demonstrated a causal relationship between neutrophils and metastasis4,5. However, the cancer-cell-intrinsic mechanisms that dictate the substantial heterogeneity in systemic neutrophilic inflammation between tumour-bearing hosts are largely unresolved. Here, using a panel of 16 distinct genetically engineered mouse models for breast cancer, we uncover a role for cancer-cell-intrinsic p53 as a key regulator of pro-metastatic neutrophils. Mechanistically, loss of p53 in cancer cells induced the secretion of WNT ligands that stimulate tumour-associated macrophages to produce IL-1β, thus driving systemic inflammation. Pharmacological and genetic blockade of WNT secretion in p53-null cancer cells reverses macrophage production of IL-1β and subsequent neutrophilic inflammation, resulting in reduced metastasis formation. Collectively, we demonstrate a mechanistic link between the loss of p53 in cancer cells, secretion of WNT ligands and systemic neutrophilia that potentiates metastatic progression. These insights illustrate the importance of the genetic makeup of breast tumours in dictating pro-metastatic systemic inflammation, and set the stage for personalized immune intervention strategies for patients with cancer.
Conflict of interest statement
M.D.W., S.B.C., D.E.M.D., M.H.v.M., M.S., I.d.R., L.H., S.M.K., S.P., C-S.H. K.V., A.P.D., R.d.K-G., E.S. I.v.d.H., W.Z. and J.J. report no competing interests. L.F.A.W. reports research funding from Genmab. T.N.S. is a consultant for Adaptive Biotechnologies, AIMM Therapeutics, Allogene Therapeutics, Amgen, Merus, Neon Therapeutics, Scenic Biotech, Third Rock Ventures, reports research support from Merck, Bristol-Myers Squibb, Merck KGaA, and is stockholder in AIMM Therapeutics, Allogene Therapeutics, Merus, Neogene Therapeutics, Neon Therapeutics, Scenic Biotech, all outside the scope of this work. K.E.d.V. reports research funding from Roche and is consultant for Third Rock Ventures, outside the scope of this work.
Figures




References
-
- Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–503. - PubMed
-
- Templeton AJ, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106 dju124. - PubMed
-
- Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016;16:431–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

