Exome sequencing of Finnish isolates enhances rare-variant association power
- PMID: 31367044
- PMCID: PMC6697530
- DOI: 10.1038/s41586-019-1457-z
Exome sequencing of Finnish isolates enhances rare-variant association power
Erratum in
-
Author Correction: Exome sequencing of Finnish isolates enhances rare-variant association power.Nature. 2019 Nov;575(7783):E4. doi: 10.1038/s41586-019-1726-x. Nature. 2019. PMID: 31686056
Abstract
Exome-sequencing studies have generally been underpowered to identify deleterious alleles with a large effect on complex traits as such alleles are mostly rare. Because the population of northern and eastern Finland has expanded considerably and in isolation following a series of bottlenecks, individuals of these populations have numerous deleterious alleles at a relatively high frequency. Here, using exome sequencing of nearly 20,000 individuals from these regions, we investigate the role of rare coding variants in clinically relevant quantitative cardiometabolic traits. Exome-wide association studies for 64 quantitative traits identified 26 newly associated deleterious alleles. Of these 26 alleles, 19 are either unique to or more than 20 times more frequent in Finnish individuals than in other Europeans and show geographical clustering comparable to Mendelian disease mutations that are characteristic of the Finnish population. We estimate that sequencing studies of populations without this unique history would require hundreds of thousands to millions of participants to achieve comparable association power.
Conflict of interest statement
Competing interests statements:
VS has participated in a conference trip sponsored by Novo Nordisk and received a honorarium from the same source for participating in an advisory board meeting. He also has ongoing research collaboration with Bayer Ltd.
HL is a member of the Nordic Expert group unconditionally supported by Gedeon Richter Nordics and has received an honorarium from Orion.
Figures


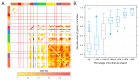


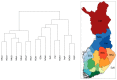

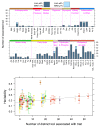

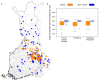
References
-
- Samocha KE, et al. Regional missense constraint improves variant deleteriousness prediction. bioRxiv. 2017 doi: 10.1101/148353. - DOI
Publication types
MeSH terms
Substances
Grants and funding
- R56 DK062370/DK/NIDDK NIH HHS/United States
- T32 NS048004/NS/NINDS NIH HHS/United States
- R01 DK062370/DK/NIDDK NIH HHS/United States
- UM1 HG008853/HG/NHGRI NIH HHS/United States
- MC_UU_12013/1/MRC_/Medical Research Council/United Kingdom
- R01 HL113315/HL/NHLBI NIH HHS/United States
- U01 MH105578/MH/NIMH NIH HHS/United States
- R01 HG006695/HG/NHGRI NIH HHS/United States
- MR/S019669/1/MRC_/Medical Research Council/United Kingdom
- P30 DK056341/DK/NIDDK NIH HHS/United States
- P30 NS062691/NS/NINDS NIH HHS/United States
- U01 DK062370/DK/NIDDK NIH HHS/United States
- R01 HL131961/HL/NHLBI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States

