Whole genome sequencing revealed microbiome in lung adenocarcinomas presented as ground-glass nodules
- PMID: 31367537
- PMCID: PMC6626866
- DOI: 10.21037/tlcr.2019.06.11
Whole genome sequencing revealed microbiome in lung adenocarcinomas presented as ground-glass nodules
Abstract
Background: Emerging evidence has suggested that dysbiosis of the microbiota may play vital roles in tumorigenesis. However, the interplay between the microbiome and lung cancer remains undetermined. In this study, we characterize the microbiome in the early stage of lung cancer, which presented as ground-glass nodules (GGNs).
Methods: We sequenced the whole genomes from 10 GGN lesions and 5 adjacent normal lung tissue samples. After being filtered with human genome sequences, the sequence reads were mapped to prokaryotic genomes refSeq and non-redundant protein database for taxa and gene functions profiling, respectively.
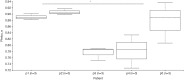
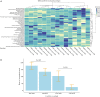
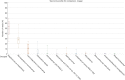
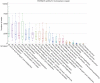
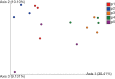
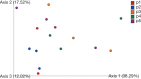
Results: Mycobacterium, Corynebacterium, and Negativicoccus were the core microbiota found in all GGNs and the normal tissue samples. The microbiota composition did not show significant difference between GGNs and normal tissues except the adenocarcinoma (AD) (P=0.047). A significant β diversity in microbiome gene functions was found among different patients. Two individual gene functions, the Secondary Metabolism (1.32 fold with P=0.01) and the Serine Threonine protein kinase (4.23 fold, P<0.001), were significantly increased in GGNs over normal tissue samples.
Conclusions: This study helps shed light on the implication of the microbiome in early stage lung cancer, which encourages the further study and development of innovative strategies for early prevention and treatment of lung cancer.
Keywords: Lung cancer; ground-glass nodule (GGN); microbiome; whole genome sequencing (WGS).
Conflict of interest statement
Conflicts of Interest: The authors have no conflicts of interest to declare.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
