Shared Pathogenomic Patterns Characterize a New Phylotype, Revealing Transition toward Host-Adaptation Long before Speciation of Mycobacterium tuberculosis
- PMID: 31368488
- PMCID: PMC6736058
- DOI: 10.1093/gbe/evz162
Shared Pathogenomic Patterns Characterize a New Phylotype, Revealing Transition toward Host-Adaptation Long before Speciation of Mycobacterium tuberculosis
Abstract
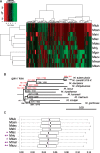
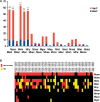
Tuberculosis remains one of the deadliest infectious diseases of humanity. To better understand the evolutionary history of host-adaptation of tubercle bacilli (MTB), we sought for mycobacterial species that were more closely related to MTB than the previously used comparator species Mycobacterium marinum and Mycobacterium kansasii. Our phylogenomic approach revealed some recently sequenced opportunistic mycobacterial pathogens, Mycobacterium decipiens, Mycobacterium lacus, Mycobacterium riyadhense, and Mycobacterium shinjukuense, to constitute a common clade with MTB, hereafter called MTB-associated phylotype (MTBAP), from which MTB have emerged. Multivariate and clustering analyses of genomic functional content revealed that the MTBAP lineage forms a clearly distinct cluster of species that share common genomic characteristics, such as loss of core genes, shift in dN/dS ratios, and massive expansion of toxin-antitoxin systems. Consistently, analysis of predicted horizontal gene transfer regions suggests that putative functions acquired by MTBAP members were markedly associated with changes in microbial ecology, for example adaption to intracellular stress resistance. Our study thus considerably deepens our view on MTB evolutionary history, unveiling a decisive shift that promoted conversion to host-adaptation among ancestral founders of the MTBAP lineage long before Mycobacterium tuberculosis has adapted to the human host.
Keywords: evolutionary transition; host adaption; mycobacteria; pathogen evolution; pathogenomic; tuberculosis.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. - PubMed
-
- Ankenbrand MJ, Keller A.. 2016. bcgTree: automatized phylogenetic tree building from bacterial core genomes. Genome 59(10):783–791. - PubMed
-
- Ates LS, Brosch R.. 2017. Discovery of the type VII ESX-1 secretion needle? Mol Microbiol. 103(1):7–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

