Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer
- PMID: 31369045
- PMCID: PMC6681568
- DOI: 10.1001/jamaoncol.2019.1838
Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer
Erratum in
-
Errors in Results.JAMA Oncol. 2020 Jan 1;6(1):162. doi: 10.1001/jamaoncol.2019.6325. JAMA Oncol. 2020. PMID: 31917389 Free PMC article. No abstract available.
Abstract
Importance: Current treatment cures most cases of early-stage, primary breast cancer. However, better techniques are required to identify which patients are at risk of relapse.
Objective: To assess the clinical validity of molecular relapse detection with circulating tumor DNA (ctDNA) analysis in early-stage breast cancer.
Design, setting, and participants: This prospective, multicenter, sample collection, validation study conducted at 5 United Kingdom medical centers from November 24, 2011, to October 18, 2016, assessed patients with early-stage breast cancer irrespective of hormone receptor and ERBB2 (formerly HER2 or HER2/neu) status who were receiving neoadjuvant chemotherapy followed by surgery or surgery before adjuvant chemotherapy. The study recruited 170 women, with mutations identified in 101 patients forming the main cohort. Secondary analyses were conducted on a combined cohort of 144 patients, including 43 patients previously analyzed in a proof of principle study.
Interventions: Primary tumor was sequenced to identify somatic mutations, and personalized tumor-specific digital polymerase chain reaction assays were used to monitor these mutations in serial plasma samples taken every 3 months for the first year of follow-up and subsequently every 6 months.
Main outcomes and measures: The primary end point was relapse-free survival analyzed with Cox proportional hazards regression models.
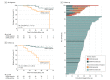
Results: In the main cohort of 101 female patients (mean [SD] age, 54 [11] years) with a median follow-up of 35.5 months (interquartile range, 27.9-43.0 months), detection of ctDNA during follow-up was associated with relapse (hazard ratio, 25.2; 95% CI, 6.7-95.6; P < .001). Detection of ctDNA at diagnosis, before any treatment, was also associated with relapse-free survival (hazard ratio, 5.8; 95% CI, 1.2-27.1; P = .01). In the combined cohort, ctDNA detection had a median lead time of 10.7 months (95% CI, 8.1-19.1 months) compared with clinical relapse and was associated with relapse in all breast cancer subtypes. Distant extracranial metastatic relapse was detected by ctDNA in 22 of 23 patients (96%). Brain-only metastasis was less commonly detected by ctDNA (1 of 6 patients [17%]), suggesting relapse sites less readily detectable by ctDNA analysis.
Conclusions and relevance: The findings suggest that detection of ctDNA during follow-up is associated with a high risk of future relapse of early-stage breast cancer. Prospective studies are needed to assess the potential of molecular relapse detection to guide adjuvant therapy.
Conflict of interest statement
Figures

Comment in
-
Clinical Benefit of Circulating Tumor DNA Analysis in Early-Stage Breast Cancer.JAMA Oncol. 2020 Mar 1;6(3):439. doi: 10.1001/jamaoncol.2019.5677. JAMA Oncol. 2020. PMID: 31855227 No abstract available.
-
Clinical Benefit of Circulating Tumor DNA Analysis in Follow-up of Patients With Early-Stage Breast Cancer-Reply.JAMA Oncol. 2020 Mar 1;6(3):439-440. doi: 10.1001/jamaoncol.2019.5682. JAMA Oncol. 2020. PMID: 31855255 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

