Comparative Proteomics and Physiological Analyses Reveal Important Maize Filling-Kernel Drought-Responsive Genes and Metabolic Pathways
- PMID: 31370198
- PMCID: PMC6696053
- DOI: 10.3390/ijms20153743
Comparative Proteomics and Physiological Analyses Reveal Important Maize Filling-Kernel Drought-Responsive Genes and Metabolic Pathways
Abstract
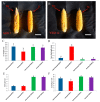
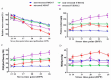
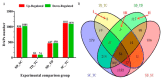
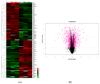
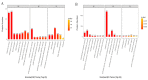
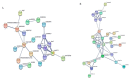
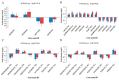
Despite recent scientific headway in deciphering maize (Zea mays L.) drought stress responses, the overall picture of key proteins and genes, pathways, and protein-protein interactions regulating maize filling-kernel drought tolerance is still fragmented. Yet, maize filling-kernel drought stress remains devastating and its study is critical for tolerance breeding. Here, through a comprehensive comparative proteomics analysis of filling-kernel proteomes of two contrasting (drought-tolerant YE8112 and drought-sensitive MO17) inbred lines, we report diverse but key molecular actors mediating drought tolerance in maize. Using isobaric tags for relative quantification approach, a total of 5175 differentially abundant proteins (DAPs) were identified from four experimental comparisons. By way of Venn diagram analysis, four critical sets of drought-responsive proteins were mined out and further analyzed by bioinformatics techniques. The YE8112-exclusive DAPs chiefly participated in pathways related to "protein processing in the endoplasmic reticulum" and "tryptophan metabolism", whereas MO17-exclusive DAPs were involved in "starch and sucrose metabolism" and "oxidative phosphorylation" pathways. Most notably, we report that YE8112 kernels were comparatively drought tolerant to MO17 kernels attributable to their redox post translational modifications and epigenetic regulation mechanisms, elevated expression of heat shock proteins, enriched energy metabolism and secondary metabolites biosynthesis, and up-regulated expression of seed storage proteins. Further, comparative physiological analysis and quantitative real time polymerase chain reaction results substantiated the proteomics findings. Our study presents an elaborate understanding of drought-responsive proteins and metabolic pathways mediating maize filling-kernel drought tolerance, and provides important candidate genes for subsequent functional validation.
Keywords: Zea mays L.; drought stress; filling kernel; heat shock proteins; iTRAQ; proteomes.
Conflict of interest statement
The authors declare that they have no conflict of interest. Additionally, the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Comparative transcriptomic analysis of contrasting hybrid cultivars reveal key drought-responsive genes and metabolic pathways regulating drought stress tolerance in maize at various stages.PLoS One. 2020 Oct 15;15(10):e0240468. doi: 10.1371/journal.pone.0240468. eCollection 2020. PLoS One. 2020. PMID: 33057352 Free PMC article.
-
Comparative Proteomic and Physiological Analyses of Two Divergent Maize Inbred Lines Provide More Insights into Drought-Stress Tolerance Mechanisms.Int J Mol Sci. 2018 Oct 18;19(10):3225. doi: 10.3390/ijms19103225. Int J Mol Sci. 2018. PMID: 30340410 Free PMC article.
-
Key Maize Drought-Responsive Genes and Pathways Revealed by Comparative Transcriptome and Physiological Analyses of Contrasting Inbred Lines.Int J Mol Sci. 2019 Mar 13;20(6):1268. doi: 10.3390/ijms20061268. Int J Mol Sci. 2019. PMID: 30871211 Free PMC article.
-
Transcriptional regulatory networks in response to drought stress and rewatering in maize (Zea mays L.).Mol Genet Genomics. 2021 Nov;296(6):1203-1219. doi: 10.1007/s00438-021-01820-y. Epub 2021 Oct 3. Mol Genet Genomics. 2021. PMID: 34601650 Review.
-
Genetic, molecular and physiological crosstalk during drought tolerance in maize (Zea mays): pathways to resilient agriculture.Planta. 2024 Aug 28;260(4):81. doi: 10.1007/s00425-024-04517-9. Planta. 2024. PMID: 39196449 Review.
Cited by
-
iTRAQ-Based Proteomic Analysis Reveals Several Strategies to Cope with Drought Stress in Maize Seedlings.Int J Mol Sci. 2019 Nov 26;20(23):5956. doi: 10.3390/ijms20235956. Int J Mol Sci. 2019. PMID: 31779286 Free PMC article.
-
Protein and Proteome Atlas for Plants under Stresses: New Highlights and Ways for Integrated Omics in Post-Genomics Era.Int J Mol Sci. 2019 Oct 21;20(20):5222. doi: 10.3390/ijms20205222. Int J Mol Sci. 2019. PMID: 31640274 Free PMC article. Review.
-
Comparative Metabolomic Studies of Siberian Wildrye (Elymus sibiricus L.): A New Look at the Mechanism of Plant Drought Resistance.Int J Mol Sci. 2022 Dec 27;24(1):452. doi: 10.3390/ijms24010452. Int J Mol Sci. 2022. PMID: 36613896 Free PMC article.
-
2D-DIGE based proteome analysis of wheat-Thinopyrum intermedium 7XL/7DS translocation line under drought stress.BMC Genomics. 2022 May 14;23(1):369. doi: 10.1186/s12864-022-08599-1. BMC Genomics. 2022. PMID: 35568798 Free PMC article.
-
Comparative transcriptomic analysis of contrasting hybrid cultivars reveal key drought-responsive genes and metabolic pathways regulating drought stress tolerance in maize at various stages.PLoS One. 2020 Oct 15;15(10):e0240468. doi: 10.1371/journal.pone.0240468. eCollection 2020. PLoS One. 2020. PMID: 33057352 Free PMC article.
References
-
- Yang L., Jiang T., Fountain J.C., Scully B.T., Lee R.D., Kemerait R.C., Chen S., Guo B. Protein profiles reveal diverse responsive signaling pathways in kernels of two maize inbred lines with contrasting drought sensitivity. Int. J. Mol. Sci. 2014;15:18892–18918. doi: 10.3390/ijms151018892. - DOI - PMC - PubMed
-
- Farooq M., Wahid A., Kobayashi N., Fujita D., Basra S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009;29:185–212. doi: 10.1051/agro:2008021. - DOI
-
- Zhang J.Y., Cruz de Carvalho M.H., Torres-Jerez I., Kang Y., Allen S.N., Huhman D.V., Tang Y., Murray J., Sumner L.W., Udvardi M.K. Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant Cell Environ. 2014;37:2553–2576. doi: 10.1111/pce.12328. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

