The prognostic value of JUNB-positive CTCs in metastatic breast cancer: from bioinformatics to phenotypic characterization
- PMID: 31370904
- PMCID: PMC6676640
- DOI: 10.1186/s13058-019-1166-4
The prognostic value of JUNB-positive CTCs in metastatic breast cancer: from bioinformatics to phenotypic characterization
Abstract
Background: Circulating tumor cells (CTCs) are important for metastatic dissemination of cancer. They can provide useful information, regarding biological features and tumor heterogeneity; however, their detection and characterization are difficult due to their limited number in the bloodstream and their mesenchymal characteristics. Therefore, new biomarkers are needed to address these questions.
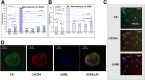
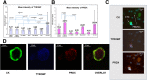
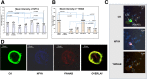
Methods: Bioinformatics functional enrichment analysis revealed a subgroup of 24 genes, potentially overexpressed in CTCs. Among these genes, the chemokine receptor CXCR4 plays a central role. After prioritization according to the CXCR4 corresponding pathways, five molecules (JUNB, YWHAB, TYROBP, NFYA, and PRDX1) were selected for further analysis in biological samples. The SKBR3, MDA-MB231, and MCF7 cell lines, as well as PBMCs from normal (n = 10) blood donors, were used as controls to define the expression pattern of all the examined molecules. Consequently, 100 previously untreated metastatic breast cancer (mBC) patients (n = 100) were analyzed using the following combinations of antibodies: CK (cytokeratin)/CXCR4/JUNB, CK/NFYA/ΥWHΑΒ (14-3-3), and CK/TYROBP/PRDX1. A threshold value for every molecule was considered the mean expression in normal PBMCs.
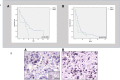
Results: Quantification of CXCR4 revealed overexpression of the receptor in SKBR3 and in CTCs, following the subsequent scale (SKBR3>CTCs>Hela>MCF7>MDA-MB231). JUNB was also overexpressed in CTCs (SKBR3>CTCs>MCF7>MDA-MB231>Hela). According to the defined threshold for each molecule, CXCR4-positive CTCs were identified in 90% of the patients with detectable tumor cells in their blood. In addition, 65%, 75%, 14.3%, and 12.5% of the patients harbored JUNB-, TYROBP-, NFYA-, and PRDX-positive CTCs, respectively. Conversely, none of the patients revealed YWHAB-positive CTCs. Interestingly, JUNB expression in CTCs was phenotypically and statistically enhanced compared to patients' blood cells (p = 0.002) providing a possible new biomarker for CTCs. Furthermore, the detection of JUNB-positive CTCs in patients was associated with poorer PFS (p = 0.015) and OS (p = 0.002). Moreover, JUNB staining of 11 primary and 4 metastatic tumors from the same cohort of patients revealed a dramatic increase of JUNB expression in metastasis.
Conclusions: CXCR4, JUNB, and TYROBP were overexpressed in CTCs, but only the expression of JUNB was associated with poor prognosis, providing a new biomarker and a potential therapeutic target for the elimination of CTCs.
Keywords: Bioinformatics; Breast cancer; CTCs; CXCR4; JUNB.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Comprehensive Analysis of CXCR4, JUNB, and PD-L1 Expression in Circulating Tumor Cells (CTCs) from Prostate Cancer Patients.Cells. 2024 May 3;13(9):782. doi: 10.3390/cells13090782. Cells. 2024. PMID: 38727318 Free PMC article.
-
CXCR4 and JUNB double-positive disseminated tumor cells are detected frequently in breast cancer patients at primary diagnosis.Ther Adv Med Oncol. 2020 Apr 28;12:1758835919895754. doi: 10.1177/1758835919895754. eCollection 2020. Ther Adv Med Oncol. 2020. PMID: 32426042 Free PMC article.
-
Phenotypic Characterization of Circulating Tumor Cells Isolated from Non-Small and Small Cell Lung Cancer Patients.Cancers (Basel). 2022 Dec 28;15(1):171. doi: 10.3390/cancers15010171. Cancers (Basel). 2022. PMID: 36612166 Free PMC article.
-
CTCs in metastatic breast cancer.Recent Results Cancer Res. 2012;195:193-201. doi: 10.1007/978-3-642-28160-0_18. Recent Results Cancer Res. 2012. PMID: 22527507 Review.
-
Profiling of Invasive Breast Carcinoma Circulating Tumour Cells-Are We Ready for the 'Liquid' Revolution?Cancers (Basel). 2019 Jan 25;11(2):143. doi: 10.3390/cancers11020143. Cancers (Basel). 2019. PMID: 30691008 Free PMC article. Review.
Cited by
-
STIM1, ORAI1, and KDM2B in circulating tumor cells (CTCs) isolated from prostate cancer patients.Front Cell Dev Biol. 2024 Jun 5;12:1399092. doi: 10.3389/fcell.2024.1399092. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38903530 Free PMC article.
-
Comprehensive Analysis of Purine-Metabolism-Related Gene Signature for Predicting Ovarian Cancer Prognosis, Immune Landscape, and Potential Treatment Options.J Pers Med. 2023 Apr 29;13(5):776. doi: 10.3390/jpm13050776. J Pers Med. 2023. PMID: 37240946 Free PMC article.
-
Integrative analysis of circulating tumor cells (CTCs) and exosomes from small-cell lung cancer (SCLC) patients: a comprehensive approach.Mol Oncol. 2025 Jul;19(7):2038-2055. doi: 10.1002/1878-0261.13765. Epub 2024 Nov 22. Mol Oncol. 2025. PMID: 39575761 Free PMC article.
-
Cancer-Associated Fibroblasts Facilitate Squamous Cell Carcinoma Lung Metastasis in Mice by Providing TGFβ-Mediated Cancer Stem Cell Niche.Front Cell Dev Biol. 2021 Aug 30;9:668164. doi: 10.3389/fcell.2021.668164. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34527666 Free PMC article.
-
Identification of Immune-Related Markers in Hepatocellular Carcinoma Based on Gene Co-expression Network.Biochem Genet. 2022 Dec;60(6):2552-2569. doi: 10.1007/s10528-022-10235-2. Epub 2022 May 28. Biochem Genet. 2022. PMID: 35633444
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

