Adaptation by Loss of Heterozygosity in Saccharomyces cerevisiae Clones Under Divergent Selection
- PMID: 31371407
- PMCID: PMC6781901
- DOI: 10.1534/genetics.119.302411
Adaptation by Loss of Heterozygosity in Saccharomyces cerevisiae Clones Under Divergent Selection
Abstract
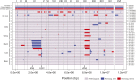
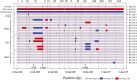
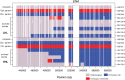
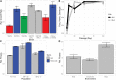
Loss of heterozygosity (LOH) is observed during vegetative growth and reproduction of diploid genotypes through mitotic crossovers, aneuploidy caused by nondisjunction, and gene conversion. We aimed to test the role that LOH plays during adaptation of two highly heterozygous Saccharomyces cerevisiae genotypes to multiple environments over a short time span in the laboratory. We hypothesized that adaptation would be observed through parallel LOH events across replicate populations. Using genome resequencing of 70 clones, we found that LOH was widespread with 5.2 LOH events per clone after ∼500 generations. The most common mode of LOH was gene conversion (51%) followed by crossing over consistent with either break-induced replication or double Holliday junction resolution. There was no evidence that LOH involved nondisjunction of whole chromosomes. We observed parallel LOH in both an environment-specific and environment-independent manner. LOH largely involved recombining existing variation between the parental genotypes, but also was observed after de novo, presumably beneficial, mutations occurred in the presence of canavanine, a toxic analog of arginine. One highly parallel LOH event involved the ENA salt efflux pump locus on chromosome IV, which showed repeated LOH to the allele from the European parent, an allele originally derived by introgression from S. paradoxus Using CRISPR-engineered LOH we showed that the fitness advantage provided by this single LOH event was 27%. Overall, we found extensive evidence that LOH could be adaptive and is likely to be a greater source of initial variation than de novo mutation for rapid evolution of diploid genotypes.
Keywords: Saccharomyces cerevisiae; experimental evolution; gene conversion; mitotic recombination; resequencing.
Copyright © 2019 by the Genetics Society of America.
Figures




References
-
- Auguié, B., 2017 gridExtra: miscellaneous functions for “Grid” graphics, R package version 2.3. https://CRAN.R-project.org/package=gridExtra.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

