Tmem119-EGFP and Tmem119-CreERT2 Transgenic Mice for Labeling and Manipulating Microglia
- PMID: 31371457
- PMCID: PMC6712208
- DOI: 10.1523/ENEURO.0448-18.2019
Tmem119-EGFP and Tmem119-CreERT2 Transgenic Mice for Labeling and Manipulating Microglia
Abstract
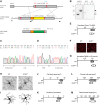
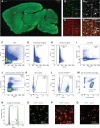
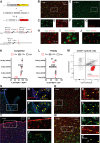
Microglia are specialized brain-resident macrophages with important functions in health and disease. To improve our understanding of these cells, the research community needs genetic tools to identify and control them in a manner that distinguishes them from closely related cell types. We have targeted the recently discovered microglia-specific Tmem119 gene to generate knock-in mice expressing EGFP (JAX#031823) or CreERT2 (JAX#031820) for the identification and manipulation of microglia, respectively. Genetic characterization of the locus and qPCR-based analysis demonstrate correct positioning of the transgenes and intact expression of endogenous Tmem119 in the knock-in mouse models. Immunofluorescence analysis further shows that parenchymal microglia, but not other brain macrophages, are completely and faithfully labeled in the EGFP-line at different time points of development. Flow cytometry indicates highly selective expression of EGFP in CD11b+CD45lo microglia. Similarly, immunofluorescence and flow cytometry analyses using a Cre-dependent reporter mouse line demonstrate activity of CreERT2 primarily in microglia upon tamoxifen administration with the caveat of activity in leptomeningeal cells. Finally, flow cytometric analyses reveal absence of EGFP expression and minimal activity of CreERT2 in blood monocytes of the Tmem119-EGFP and Tmem119-CreERT2 lines, respectively. These new transgenic lines extend the microglia toolbox by providing the currently most specific genetic labeling and control over these cells in the myeloid compartment of mice.
Keywords: CreERT2; EGFP; Tmem119; macrophage; microglia; transgenic.
Copyright © 2019 Kaiser and Feng.
Figures




References
-
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, Weissman IL, Chang EF, Li G, Grant GA, Hayden Gephart MG, Barres BA (2016) New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113:E1738–E1746. 10.1073/pnas.1525528113 - DOI - PMC - PubMed
-
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL (2014) Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat Neurosci 17:131. 10.1038/nn.3599 - DOI - PMC - PubMed
-
- Clausen B, Burkhardt C, Reith W, Renkawitz R, Förster I (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8:265–277. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
