Myosin V regulates synaptopodin clustering and localization in the dendrites of hippocampal neurons
- PMID: 31371487
- PMCID: PMC6737913
- DOI: 10.1242/jcs.230177
Myosin V regulates synaptopodin clustering and localization in the dendrites of hippocampal neurons
Abstract
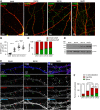
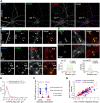
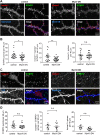
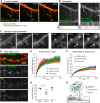
The spine apparatus (SA) is an endoplasmic reticulum-related organelle that is present in a subset of dendritic spines in cortical and pyramidal neurons, and plays an important role in Ca2+ homeostasis and dendritic spine plasticity. The protein synaptopodin is essential for the formation of the SA and is widely used as a maker for this organelle. However, it is still unclear which factors contribute to its localization at selected synapses, and how it triggers local SA formation. In this study, we characterized development, localization and mobility of synaptopodin clusters in hippocampal primary neurons, as well as the molecular dynamics within these clusters. Interestingly, synaptopodin at the shaft-associated clusters is less dynamic than at spinous clusters. We identify the actin-based motor proteins myosin V (herein referring to both the myosin Va and Vb forms) and VI as novel interaction partners of synaptopodin, and demonstrate that myosin V is important for the formation and/or maintenance of the SA. We found no evidence of active microtubule-based transport of synaptopodin. Instead, new clusters emerge inside spines, which we interpret as the SA being assembled on-site.
Keywords: Dendritic spines; F-actin; Myosin; Spine apparatus; Synaptopodin.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


Similar articles
-
Caldendrin and myosin V regulate synaptic spine apparatus localization via ER stabilization in dendritic spines.EMBO J. 2022 Feb 15;41(4):e106523. doi: 10.15252/embj.2020106523. Epub 2021 Dec 22. EMBO J. 2022. PMID: 34935159 Free PMC article.
-
Actin-associated protein synaptopodin in the rat hippocampal formation: localization in the spine neck and close association with the spine apparatus of principal neurons.J Comp Neurol. 2000 Mar 6;418(2):164-81. doi: 10.1002/(sici)1096-9861(20000306)418:2<164::aid-cne4>3.0.co;2-0. J Comp Neurol. 2000. PMID: 10701442
-
Lamina-specific distribution of Synaptopodin, an actin-associated molecule essential for the spine apparatus, in identified principal cell dendrites of the mouse hippocampus.J Comp Neurol. 2005 Jul 4;487(3):227-39. doi: 10.1002/cne.20539. J Comp Neurol. 2005. PMID: 15892100
-
Potential role of synaptopodin in spine motility by coupling actin to the spine apparatus.Hippocampus. 2000;10(5):569-81. doi: 10.1002/1098-1063(2000)10:5<569::AID-HIPO7>3.0.CO;2-M. Hippocampus. 2000. PMID: 11075827 Review.
-
Understanding the role of synaptopodin and the spine apparatus in Hebbian synaptic plasticity - New perspectives and the need for computational modeling.Neurobiol Learn Mem. 2017 Feb;138:21-30. doi: 10.1016/j.nlm.2016.07.023. Epub 2016 Jul 25. Neurobiol Learn Mem. 2017. PMID: 27470091 Review.
Cited by
-
Proximity proteomics of synaptopodin provides insight into the molecular composition of the spine apparatus of dendritic spines.Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2203750119. doi: 10.1073/pnas.2203750119. Epub 2022 Oct 10. Proc Natl Acad Sci U S A. 2022. PMID: 36215465 Free PMC article.
-
High levels of 27-hydroxycholesterol results in synaptic plasticity alterations in the hippocampus.Sci Rep. 2021 Feb 12;11(1):3736. doi: 10.1038/s41598-021-83008-3. Sci Rep. 2021. PMID: 33580102 Free PMC article.
-
The actin-modulating protein synaptopodin mediates long-term survival of dendritic spines.Elife. 2020 Dec 4;9:e62944. doi: 10.7554/eLife.62944. Elife. 2020. PMID: 33275099 Free PMC article.
-
A novel labeling strategy reveals that myosin Va and myosin Vb bind the same dendritically polarized vesicle population.Traffic. 2020 Nov;21(11):689-701. doi: 10.1111/tra.12764. Traffic. 2020. PMID: 32959500 Free PMC article.
-
Caldendrin and myosin V regulate synaptic spine apparatus localization via ER stabilization in dendritic spines.EMBO J. 2022 Feb 15;41(4):e106523. doi: 10.15252/embj.2020106523. Epub 2021 Dec 22. EMBO J. 2022. PMID: 34935159 Free PMC article.
References
-
- Avraham K. B., Hasson T., Steel K. P., Kingsley D. M., Russel L. B., Mooseker M. S., Copeland N. G. and Jenkins N. A. (1995). The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nature 11, 369-375. 10.1038/ng1295-369 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

