Subunit interactions and arrangements in the fission yeast Mis16-Mis18-Mis19 complex
- PMID: 31371524
- PMCID: PMC6677171
- DOI: 10.26508/lsa.201900408
Subunit interactions and arrangements in the fission yeast Mis16-Mis18-Mis19 complex
Abstract
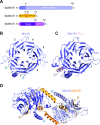
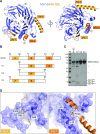
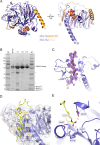
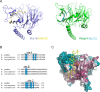
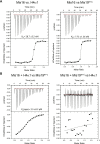
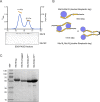
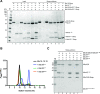
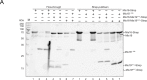
Centromeric chromatin in fission yeast is distinguished by the presence of nucleosomes containing the histone H3 variant Cnp1CENP-A Cell cycle-specific deposition of Cnp1 requires the Mis16-Mis18-Mis19 complex, which is thought to direct recruitment of Scm3-chaperoned Cnp1/histone H4 dimers to DNA. Here, we present the structure of the essential Mis18 partner protein Mis19 and describe its interaction with Mis16, revealing a bipartite-binding site. We provide data on the stoichiometry and overall architecture of the complex and provide detailed insights into the Mis18-Mis19 interface.
© 2019 Korntner-Vetter et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. 10.1107/s0907444909052925 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
