Expansion of a core regulon by transposable elements promotes Arabidopsis chemical diversity and pathogen defense
- PMID: 31371717
- PMCID: PMC6671987
- DOI: 10.1038/s41467-019-11406-3
Expansion of a core regulon by transposable elements promotes Arabidopsis chemical diversity and pathogen defense
Abstract
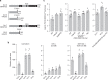
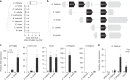
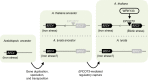
Plants synthesize numerous ecologically specialized, lineage-specific metabolites through biosynthetic gene duplication and functional specialization. However, it remains unclear how duplicated genes are wired into existing regulatory networks. We show that the duplicated gene CYP82C2 has been recruited into the WRKY33 regulon and indole-3-carbonylnitrile (ICN) biosynthetic pathway through exaptation of a retroduplicated LINE retrotransposon (EPCOT3) into an enhancer. The stepwise development of a chromatin-accessible WRKY33-binding site on EPCOT3 has potentiated the regulatory neofunctionalization of CYP82C2 and the evolution of inducible defense metabolite 4-hydroxy-ICN in Arabidopsis thaliana. Although transposable elements (TEs) have long been recognized to have the potential to rewire regulatory networks, these results establish a more complete understanding of how duplicated genes and TEs contribute in concert to chemical diversity and pathogen defense.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Ohno, S. Evolution by Gene Duplication (Springer-Verlag, Berlin, 1970).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

