A family of anti-Bacteroidales peptide toxins wide-spread in the human gut microbiota
- PMID: 31371723
- PMCID: PMC6671954
- DOI: 10.1038/s41467-019-11494-1
A family of anti-Bacteroidales peptide toxins wide-spread in the human gut microbiota
Abstract
Bacteria often produce antimicrobial toxins to compete in microbial communities. Here we identify a family of broad-spectrum peptide toxins, named bacteroidetocins, produced by Bacteroidetes species. We study this toxin family using phenotypic, mutational, bioinformatic, and human metagenomic analyses. Bacteroidetocins are related to class IIa bacteriocins of Gram-positive bacteria and kill members of the Bacteroidetes phylum, including Bacteroides, Parabacteroides, and Prevotella gut species, as well as pathogenic Prevotella species. The bacteroidetocin biosynthesis genes are found in horizontally acquired mobile elements, which likely allow dissemination within the gut microbiota and may explain their wide distribution in human populations. Bacteroidetocins may have potential applications in microbiome engineering and as therapeutics for polymicrobial diseases such as bacterial vaginosis and periodontal disease.
Conflict of interest statement
The authors declare no competing interests.
Figures



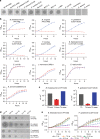
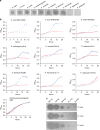
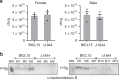


Similar articles
-
Bacteroidetocins Target the Essential Outer Membrane Protein BamA of Bacteroidales Symbionts and Pathogens.mBio. 2021 Oct 26;12(5):e0228521. doi: 10.1128/mBio.02285-21. Epub 2021 Sep 14. mBio. 2021. PMID: 34517753 Free PMC article.
-
Streamlined Genetic Manipulation of Diverse Bacteroides and Parabacteroides Isolates from the Human Gut Microbiota.mBio. 2019 Aug 13;10(4):e01762-19. doi: 10.1128/mBio.01762-19. mBio. 2019. PMID: 31409684 Free PMC article.
-
Evidence of extensive DNA transfer between bacteroidales species within the human gut.mBio. 2014 Jun 17;5(3):e01305-14. doi: 10.1128/mBio.01305-14. mBio. 2014. PMID: 24939888 Free PMC article.
-
Type VI Secretion Systems and the Gut Microbiota.Microbiol Spectr. 2019 Mar;7(2):10.1128/microbiolspec.psib-0009-2018. doi: 10.1128/microbiolspec.PSIB-0009-2018. Microbiol Spectr. 2019. PMID: 30825301 Free PMC article. Review.
-
Parabacteroides distasonis: intriguing aerotolerant gut anaerobe with emerging antimicrobial resistance and pathogenic and probiotic roles in human health.Gut Microbes. 2021 Jan-Dec;13(1):1922241. doi: 10.1080/19490976.2021.1922241. Gut Microbes. 2021. PMID: 34196581 Free PMC article. Review.
Cited by
-
Animal Microbiomes as a Source of Novel Antibiotic-Producing Strains.Int J Mol Sci. 2023 Dec 30;25(1):537. doi: 10.3390/ijms25010537. Int J Mol Sci. 2023. PMID: 38203702 Free PMC article. Review.
-
High biodiversity in a benzene-degrading nitrate-reducing culture is sustained by a few primary consumers.Commun Biol. 2021 May 5;4(1):530. doi: 10.1038/s42003-021-01948-y. Commun Biol. 2021. PMID: 33953314 Free PMC article.
-
A proteolytically activated antimicrobial toxin encoded on a mobile plasmid of Bacteroidales induces a protective response.Nat Commun. 2022 Jul 23;13(1):4258. doi: 10.1038/s41467-022-31925-w. Nat Commun. 2022. PMID: 35871068 Free PMC article.
-
Genetic and Biochemical Analysis of Anaerobic Respiration in Bacteroides fragilis and Its Importance In Vivo.mBio. 2020 Feb 4;11(1):e03238-19. doi: 10.1128/mBio.03238-19. mBio. 2020. PMID: 32019804 Free PMC article.
-
Microbiota-mediated colonization resistance: mechanisms and regulation.Nat Rev Microbiol. 2023 Jun;21(6):347-360. doi: 10.1038/s41579-022-00833-7. Epub 2022 Dec 20. Nat Rev Microbiol. 2023. PMID: 36539611 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

