Plant chemical genetics reveals colistin sulphate as a SA and NPR1-independent PR1 inducer functioning via a p38-like kinase pathway
- PMID: 31371749
- PMCID: PMC6671972
- DOI: 10.1038/s41598-019-47526-5
Plant chemical genetics reveals colistin sulphate as a SA and NPR1-independent PR1 inducer functioning via a p38-like kinase pathway
Abstract
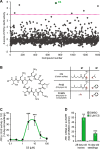
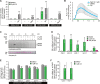
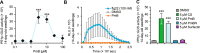
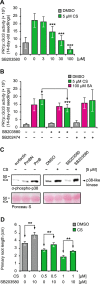
In plants, low-dose of exogenous bacterial cyclic lipopeptides (CLPs) trigger transient membrane changes leading to activation of early and late defence responses. Here, a forward chemical genetics approach identifies colistin sulphate (CS) CLP as a novel plant defence inducer. CS uniquely triggers activation of the PATHOGENESIS-RELATED 1 (PR1) gene and resistance against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) in Arabidopsis thaliana (Arabidopsis) independently of the PR1 classical inducer, salicylic acid (SA) and the key SA-signalling protein, NON-EXPRESSOR OF PR1 (NPR1). Low bioactive concentration of CS does not trigger activation of early defence markers such as reactive oxygen species (ROS) and mitogen activated protein kinase (MAPK). However, it strongly suppresses primary root length elongation. Structure activity relationship (SAR) assays and mode-of-action (MoA) studies show the acyl chain and activation of a ∼46 kDa p38-like kinase pathway to be crucial for CS' bioactivity. Selective pharmacological inhibition of the active p38-like kinase pathway by SB203580 reverses CS' effects on PR1 activation and root length suppression. Our results with CS as a chemical probe highlight the existence of a novel SA- and NPR1-independent branch of PR1 activation functioning via a membrane-sensitive p38-like kinase pathway.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae.Plant Physiol Biochem. 2013 Dec;73:383-91. doi: 10.1016/j.plaphy.2013.10.024. Epub 2013 Oct 26. Plant Physiol Biochem. 2013. PMID: 24215930
-
Priming for enhanced defence responses by specific inhibition of the Arabidopsis response to coronatine.Plant J. 2011 Feb;65(3):469-79. doi: 10.1111/j.1365-313X.2010.04436.x. Epub 2010 Dec 30. Plant J. 2011. PMID: 21265899
-
INDUCER OF CBF EXPRESSION 1 promotes cold-enhanced immunity by directly activating salicylic acid signaling.Plant Cell. 2024 Jul 2;36(7):2587-2606. doi: 10.1093/plcell/koae096. Plant Cell. 2024. PMID: 38536743 Free PMC article.
-
NPR1 in JazzSet with Pathogen Effectors.Trends Plant Sci. 2018 Jun;23(6):469-472. doi: 10.1016/j.tplants.2018.04.007. Epub 2018 May 9. Trends Plant Sci. 2018. PMID: 29753632 Review.
-
Research Progress of ATGs Involved in Plant Immunity and NPR1 Metabolism.Int J Mol Sci. 2021 Nov 9;22(22):12093. doi: 10.3390/ijms222212093. Int J Mol Sci. 2021. PMID: 34829975 Free PMC article. Review.
Cited by
-
A high-throughput differential chemical genetic screen uncovers genotype-specific compounds altering plant growth.iScience. 2025 Apr 8;28(5):112375. doi: 10.1016/j.isci.2025.112375. eCollection 2025 May 16. iScience. 2025. PMID: 40292320 Free PMC article.
-
An open source plant kinase chemogenomics set.Plant Direct. 2022 Nov 25;6(11):e460. doi: 10.1002/pld3.460. eCollection 2022 Nov. Plant Direct. 2022. PMID: 36447653 Free PMC article.
-
Overexpression of the C4 protein of tomato yellow leaf curl Sardinia virus increases tomato resistance to powdery mildew.Front Plant Sci. 2023 Mar 31;14:1163315. doi: 10.3389/fpls.2023.1163315. eCollection 2023. Front Plant Sci. 2023. PMID: 37063219 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

