Structure of Myosin VI/Tom1 complex reveals a cargo recognition mode of Myosin VI for tethering
- PMID: 31371777
- PMCID: PMC6673701
- DOI: 10.1038/s41467-019-11481-6
Structure of Myosin VI/Tom1 complex reveals a cargo recognition mode of Myosin VI for tethering
Abstract
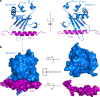
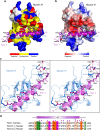
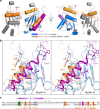
Myosin VI plays crucial roles in diverse cellular processes. In autophagy, Myosin VI can facilitate the maturation of autophagosomes through interactions with Tom1 and the autophagy receptors, Optineurin, NDP52 and TAX1BP1. Here, we report the high-resolution crystal structure of the C-terminal cargo-binding domain (CBD) of Myosin VI in complex with Tom1, which elucidates the mechanistic basis underpinning the specific interaction between Myosin VI and Tom1, and uncovers that the C-terminal CBD of Myosin VI adopts a unique cargo recognition mode to interact with Tom1 for tethering. Furthermore, we show that Myosin VI can serve as a bridging adaptor to simultaneously interact with Tom1 and autophagy receptors through two distinct interfaces. In all, our findings provide mechanistic insights into the interactions of Myosin VI with Tom1 and relevant autophagy receptors, and are valuable for further understanding the functions of these proteins in autophagy and the cargo recognition modes of Myosin VI.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome.Nat Cell Biol. 2012 Oct;14(10):1024-35. doi: 10.1038/ncb2589. Epub 2012 Sep 30. Nat Cell Biol. 2012. PMID: 23023224 Free PMC article.
-
Myosin VI and its cargo adaptors - linking endocytosis and autophagy.J Cell Sci. 2013 Jun 15;126(Pt 12):2561-70. doi: 10.1242/jcs.095554. Epub 2013 Jun 18. J Cell Sci. 2013. PMID: 23781020 Free PMC article. Review.
-
Mechanistic Insights into Recognitions of Ubiquitin and Myosin VI by Autophagy Receptor TAX1BP1.J Mol Biol. 2018 Sep 14;430(18 Pt B):3283-3296. doi: 10.1016/j.jmb.2018.06.030. Epub 2018 Jun 27. J Mol Biol. 2018. PMID: 29940186
-
Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11651-E11660. doi: 10.1073/pnas.1811421115. Epub 2018 Nov 20. Proc Natl Acad Sci U S A. 2018. PMID: 30459273 Free PMC article.
-
The MYO6 interactome: selective motor-cargo complexes for diverse cellular processes.FEBS Lett. 2019 Jul;593(13):1494-1507. doi: 10.1002/1873-3468.13486. Epub 2019 Jul 3. FEBS Lett. 2019. PMID: 31206648 Review.
Cited by
-
Mitophagy and Neurodegeneration: Between the Knowns and the Unknowns.Front Cell Dev Biol. 2022 Mar 22;10:837337. doi: 10.3389/fcell.2022.837337. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35392168 Free PMC article. Review.
-
Cardiolipin membranes drive Myosin VI activation, oligomerization, and processive cargo transport.Proc Natl Acad Sci U S A. 2025 Jun 3;122(22):e2501022122. doi: 10.1073/pnas.2501022122. Epub 2025 May 28. Proc Natl Acad Sci U S A. 2025. PMID: 40434640
-
Myosin VI maintains the actin-dependent organization of the tubulobulbar complexes required for endocytosis during mouse spermiogenesis†‡.Biol Reprod. 2020 Apr 15;102(4):863-875. doi: 10.1093/biolre/ioz232. Biol Reprod. 2020. PMID: 31901088 Free PMC article.
-
Autoinhibition and activation of myosin VI revealed by its cryo-EM structure.Nat Commun. 2024 Feb 8;15(1):1187. doi: 10.1038/s41467-024-45424-7. Nat Commun. 2024. PMID: 38331992 Free PMC article.
-
Herb pair of Astragali Radix-Descurainiae Semen attenuate heart failure through the myosin VI-Tom1 complex mediated autophagy.Front Cardiovasc Med. 2025 Jul 4;12:1599746. doi: 10.3389/fcvm.2025.1599746. eCollection 2025. Front Cardiovasc Med. 2025. PMID: 40687556 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

