IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma
- PMID: 31371825
- PMCID: PMC6686884
- DOI: 10.1038/s41556-019-0363-9
IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma
Abstract
In many cancers, high proliferation rates correlate with elevation of rRNA and tRNA levels, and nucleolar hypertrophy. However, the underlying mechanisms linking increased nucleolar transcription and tumorigenesis are only minimally understood. Here we show that IMP dehydrogenase-2 (IMPDH2), the rate-limiting enzyme for de novo guanine nucleotide biosynthesis, is overexpressed in the highly lethal brain cancer glioblastoma. This leads to increased rRNA and tRNA synthesis, stabilization of the nucleolar GTP-binding protein nucleostemin, and enlarged, malformed nucleoli. Pharmacological or genetic inactivation of IMPDH2 in glioblastoma reverses these effects and inhibits cell proliferation, whereas untransformed glia cells are unaffected by similar IMPDH2 perturbations. Impairment of IMPDH2 activity triggers nucleolar stress and growth arrest of glioblastoma cells even in the absence of functional p53. Our results reveal that upregulation of IMPDH2 is a prerequisite for the occurance of aberrant nucleolar function and increased anabolic processes in glioblastoma, which constitutes a primary event in gliomagenesis.
Conflict of interest statement
Competing interest
R.J.D. is an advisor for Agios Pharmaceuticals. P.S.M. is a cofounder, has equity and serves as a consultant for Pretzel Therapeutics, Inc.
Figures
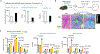




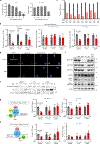

References
References for the main text
-
- Pianese G, Teuscher R & Ziegler E Beitrag zur Histologie und Aetiologie des Carcinoms. (1896).
-
- Hara A et al. Correlation between nucleolar organizer region staining and Ki-67 immunostaining in human gliomas. Surg Neurol 33, 320–324 (1990). - PubMed
-
- Busch H, Byvoet P & Smetana K The nucleolus of the cancer cell: a review. Cancer Research 23, 313–339 (1963). - PubMed
-
- Tsai RYL & Pederson T Connecting the nucleolus to the cell cycle and human disease. FASEB J. 28, 3290–3296 (2014). - PubMed
References for the Methods
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

