Accelerating structure-function mapping using the ViVa webtool to mine natural variation
- PMID: 31372596
- PMCID: PMC6658840
- DOI: 10.1002/pld3.147
Accelerating structure-function mapping using the ViVa webtool to mine natural variation
Abstract
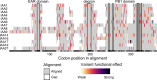
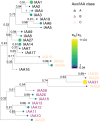
Thousands of sequenced genomes are now publicly available capturing a significant amount of natural variation within plant species; yet, much of these data remain inaccessible to researchers without significant bioinformatics experience. Here, we present a webtool called ViVa (Visualizing Variation) which aims to empower any researcher to take advantage of the amazing genetic resource collected in the Arabidopsis thaliana 1001 Genomes Project (http://1001genomes.org). ViVa facilitates data mining on the gene, gene family, or gene network level. To test the utility and accessibility of ViVa, we assembled a team with a range of expertise within biology and bioinformatics to analyze the natural variation within the well-studied nuclear auxin signaling pathway. Our analysis has provided further confirmation of existing knowledge and has also helped generate new hypotheses regarding this well-studied pathway. These results highlight how natural variation could be used to generate and test hypotheses about less-studied gene families and networks, especially when paired with biochemical and genetic characterization. ViVa is also readily extensible to databases of interspecific genetic variation in plants as well as other organisms, such as the 3,000 Rice Genomes Project ( http://snp-seek.irri.org/) and human genetic variation ( https://www.ncbi.nlm.nih.gov/clinvar/).
Keywords: Arabidopsis thaliana; accessibility; genome diversity; genotype‐phenotype; natural variation; structure‐function.
Conflict of interest statement
The authors declare no conflict of interest associated with the work described in this manuscript.
Figures




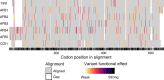



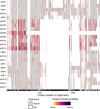
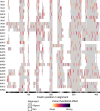
References
-
- Alexandre, C. M. , Urton, J. R. , Jean‐Baptiste, K. , Huddleston, J. , Dorrity, M. W. , Cuperus, J. T. , … Queitsch, C. (2018). Complex relationships between chromatin accessibility, sequence divergence, and gene expression in Arabidopsis thaliana . Molecular Biology and Evolution, 35(4), 837–854. 10.1093/molbev/msx326 - DOI - PMC - PubMed
-
- Allaire, J. J. , Ushey, K. , & Tang, Y. (2018). Reticulate: Interface to ‘Python’. Retrieved from https://CRAN.R-project.org/package=reticulate
-
- Allaire, J. J. , Xie, Y. , McPherson, J. , Luraschi, J. , Ushey, K. , Atkins, A. , … Iannone, R. (2018). Rmarkdown: Dynamic documents for R. Retrieved from https://CRAN.R-project.org/package=rmarkdown
-
- Aphalo, P. J. (2018a). Gginnards: Explore the innards of ‘Ggplot2’ objects. Retrieved from https://CRAN.R-project.org/package=gginnards
-
- Aphalo, P. J. (2018b). Ggpmisc: Miscellaneous extensions to ‘Ggplot2’. Retrieved from https://CRAN.R-project.org/package=ggpmisc
Grants and funding
LinkOut - more resources
Full Text Sources

