Predicting the effects of SNPs on transcription factor binding affinity
- PMID: 31373606
- PMCID: PMC7999143
- DOI: 10.1093/bioinformatics/btz612
Predicting the effects of SNPs on transcription factor binding affinity
Abstract
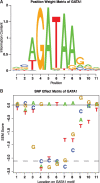
Motivation: Genome-wide association studies have revealed that 88% of disease-associated single-nucleotide polymorphisms (SNPs) reside in noncoding regions. However, noncoding SNPs remain understudied, partly because they are challenging to prioritize for experimental validation. To address this deficiency, we developed the SNP effect matrix pipeline (SEMpl).
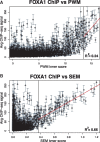
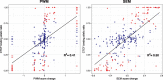
Results: SEMpl estimates transcription factor-binding affinity by observing differences in chromatin immunoprecipitation followed by deep sequencing signal intensity for SNPs within functional transcription factor-binding sites (TFBSs) genome-wide. By cataloging the effects of every possible mutation within the TFBS motif, SEMpl can predict the consequences of SNPs to transcription factor binding. This knowledge can be used to identify potential disease-causing regulatory loci.
Availability and implementation: SEMpl is available from https://github.com/Boyle-Lab/SEM_CPP.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures




References
-
- Aghera N. et al. (2011) Equilibrium unfolding studies of monellin: the double-chain variant appears to be more stable than the single-chain variant. Biochemistry, 50, 2434–2444. - PubMed
-
- Alipanahi B. et al. (2015) Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol., 33, 831–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

