Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells
- PMID: 31374860
- PMCID: PMC6721513
- DOI: 10.3390/cells8080808
Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells
Abstract
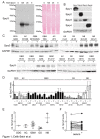
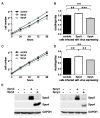
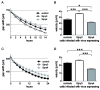
Dysregulation of receptor tyrosine kinase-induced pathways is a critical step driving the oncogenic potential of brain cancer. In this study, we investigated the role of two members of the Sprouty (Spry) family in brain cancer-derived cell lines. Using immunoblot analyses we found essential differences in the pattern of endogenous Spry3 and Spry4 expression. While Spry4 expression was mitogen-dependent and repressed in a number of cells from higher malignant brain cancers, Spry3 levels neither fluctuated in response to serum withdrawal nor were repressed in glioblastoma (GBM)-derived cell lines. In accordance to the well-known inhibitory role of Spry proteins in fibroblast growth factor (FGF)-mediated signaling, both Spry proteins were able to interfere with FGF-induced activation of the MAPK pathway although to a different extent. In response to serum solely, Spry4 exerts its role as a negative regulator of MAPK activation. Ectopic expression of Spry4 inhibited proliferation and migration of GBM-originated cells, positioning it as a tumor suppressor in brain cancer. In contrast, elevated Spry3 levels accelerated both proliferation and migration of these cell lines, while repression of Spry3 levels using shRNA caused a significant diminished growth and migration velocity rate of a GBM-derived cell line. This argues for a tumor-promoting function of Spry3 in GBMs. Based on these data we conclude that Spry3 and Spry4 fulfill different if not opposing roles within the cancerogenesis of brain malignancies.
Keywords: FGF-mediated signaling; Sprouty proteins; brain cancer; tumor promoter; tumor suppressor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Sprouty4 correlates with favorable prognosis in perihilar cholangiocarcinoma by blocking the FGFR-ERK signaling pathway and arresting the cell cycle.EBioMedicine. 2019 Dec;50:166-177. doi: 10.1016/j.ebiom.2019.11.021. Epub 2019 Nov 22. EBioMedicine. 2019. PMID: 31761616 Free PMC article.
-
A Sprouty4 Mutation Identified in Kallmann Syndrome Increases the Inhibitory Potency of the Protein towards FGF and Connected Processes.Int J Mol Sci. 2021 Feb 21;22(4):2145. doi: 10.3390/ijms22042145. Int J Mol Sci. 2021. PMID: 33670044 Free PMC article.
-
Sprouty4 mediates amphiregulin-induced down-regulation of E-cadherin and cell invasion in human ovarian cancer cells.Tumour Biol. 2016 Jul;37(7):9197-207. doi: 10.1007/s13277-016-4790-y. Epub 2016 Jan 14. Tumour Biol. 2016. PMID: 26768617
-
Fibrosis in the lens. Sprouty regulation of TGFβ-signaling prevents lens EMT leading to cataract.Exp Eye Res. 2016 Jan;142:92-101. doi: 10.1016/j.exer.2015.02.004. Epub 2015 May 21. Exp Eye Res. 2016. PMID: 26003864 Free PMC article. Review.
-
Sprouty proteins, masterminds of receptor tyrosine kinase signaling.Angiogenesis. 2008;11(1):53-62. doi: 10.1007/s10456-008-9089-1. Epub 2008 Jan 25. Angiogenesis. 2008. PMID: 18219583 Review.
Cited by
-
The roles of FGFR3 and c-MYC in urothelial bladder cancer.Discov Oncol. 2024 Jul 20;15(1):295. doi: 10.1007/s12672-024-01173-z. Discov Oncol. 2024. PMID: 39031286 Free PMC article. Review.
-
Sprouty4 correlates with favorable prognosis in perihilar cholangiocarcinoma by blocking the FGFR-ERK signaling pathway and arresting the cell cycle.EBioMedicine. 2019 Dec;50:166-177. doi: 10.1016/j.ebiom.2019.11.021. Epub 2019 Nov 22. EBioMedicine. 2019. PMID: 31761616 Free PMC article.
-
Sprouty4 is epigenetically upregulated in human colorectal cancer.Epigenetics. 2023 Dec;18(1):2145068. doi: 10.1080/15592294.2022.2145068. Epub 2022 Nov 16. Epigenetics. 2023. PMID: 36384366 Free PMC article.
-
MicroRNA and mRNA Expression Changes in Glioblastoma Cells Cultivated under Conditions of Neurosphere Formation.Curr Issues Mol Biol. 2022 Oct 30;44(11):5294-5311. doi: 10.3390/cimb44110360. Curr Issues Mol Biol. 2022. PMID: 36354672 Free PMC article.
-
LINC00675 Suppresses Cell Proliferation and Migration via Downregulating the H3K4me2 Level at the SPRY4 Promoter in Gastric Cancer.Mol Ther Nucleic Acids. 2020 Oct 4;22:766-778. doi: 10.1016/j.omtn.2020.09.038. eCollection 2020 Dec 4. Mol Ther Nucleic Acids. 2020. PMID: 33230474 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

