Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo
- PMID: 31375314
- PMCID: PMC7138101
- DOI: 10.1016/j.neuron.2019.07.002
Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo
Abstract
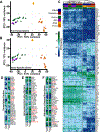
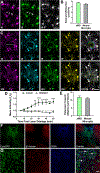
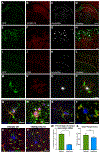
iPSC-derived microglia offer a powerful tool to study microglial homeostasis and disease-associated inflammatory responses. Yet, microglia are highly sensitive to their environment, exhibiting transcriptomic deficiencies when kept in isolation from the brain. Furthermore, species-specific genetic variations demonstrate that rodent microglia fail to fully recapitulate the human condition. To address this, we developed an approach to study human microglia within a surrogate brain environment. Transplantation of iPSC-derived hematopoietic-progenitors into the postnatal brain of humanized, immune-deficient mice results in context-dependent differentiation into microglia and other CNS macrophages, acquisition of an ex vivo human microglial gene signature, and responsiveness to both acute and chronic insults. Most notably, transplanted microglia exhibit robust transcriptional responses to Aβ-plaques that only partially overlap with that of murine microglia, revealing new, human-specific Aβ-responsive genes. We therefore have demonstrated that this chimeric model provides a powerful new system to examine the in vivo function of patient-derived and genetically modified microglia.
Keywords: Alzheimer’s disease; TREM-2; beta-amyloid; chimera; hematopoietic; humanized; microglia; neurodegeneration; pluripotent; stem cells.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Mouse brains, human microglia.Nat Rev Neurol. 2019 Oct;15(10):558-559. doi: 10.1038/s41582-019-0252-7. Nat Rev Neurol. 2019. PMID: 31462751 No abstract available.
-
Modeling neurological disease using human stem cell-derived microglia-like cells transplanted into rodent brains.Lab Anim (NY). 2020 Feb;49(2):49-51. doi: 10.1038/s41684-019-0465-9. Lab Anim (NY). 2020. PMID: 31932736 No abstract available.
References
-
- Andrews S (2014). FastQC: a quality control tool for high throughput sequence data.
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008620/GM/NIGMS NIH HHS/United States
- RF1 AG048099/AG/NIA NIH HHS/United States
- DP2 GM119164/GM/NIGMS NIH HHS/United States
- K08 NS109200/NS/NINDS NIH HHS/United States
- T32 AG000096/AG/NIA NIH HHS/United States
- R01 AG061895/AG/NIA NIH HHS/United States
- RF1 AG055524/AG/NIA NIH HHS/United States
- RF1 DA048813/DA/NIDA NIH HHS/United States
- R01 EY028212/EY/NEI NIH HHS/United States
- P30 CA062203/CA/NCI NIH HHS/United States
- F30 EY029596/EY/NEI NIH HHS/United States
- S10 OD021718/OD/NIH HHS/United States
- F31 EY028046/EY/NEI NIH HHS/United States
- T32 NS082174/NS/NINDS NIH HHS/United States
- R01 AG056303/AG/NIA NIH HHS/United States
- S10 RR025496/RR/NCRR NIH HHS/United States
- P50 AG016573/AG/NIA NIH HHS/United States
- R01 NS096170/NS/NINDS NIH HHS/United States
- S10 OD010794/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

