Class-Switch Recombination Occurs Infrequently in Germinal Centers
- PMID: 31375460
- PMCID: PMC6914312
- DOI: 10.1016/j.immuni.2019.07.001
Class-Switch Recombination Occurs Infrequently in Germinal Centers
Abstract
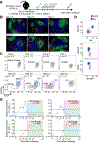
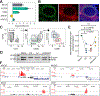
Class-switch recombination (CSR) is a DNA recombination process that replaces the immunoglobulin (Ig) constant region for the isotype that can best protect against the pathogen. Dysregulation of CSR can cause self-reactive BCRs and B cell lymphomas; understanding the timing and location of CSR is therefore important. Although CSR commences upon T cell priming, it is generally considered a hallmark of germinal centers (GCs). Here, we have used multiple approaches to show that CSR is triggered prior to differentiation into GC B cells or plasmablasts and is greatly diminished in GCs. Despite finding a small percentage of GC B cells expressing germline transcripts, phylogenetic trees of GC BCRs from secondary lymphoid organs revealed that the vast majority of CSR events occurred prior to the onset of somatic hypermutation. As such, we have demonstrated the existence of IgM-dominated GCs, which are unlikely to occur under the assumption of ongoing switching.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures




Comment in
-
Class-switch recombination revised.Nat Rev Immunol. 2019 Oct;19(10):596-597. doi: 10.1038/s41577-019-0214-8. Nat Rev Immunol. 2019. PMID: 31427740 No abstract available.
-
B Cells Hit a Class Ceiling in the Germinal Center.Immunity. 2019 Aug 20;51(2):206-208. doi: 10.1016/j.immuni.2019.07.004. Immunity. 2019. PMID: 31433966
References
-
- Al-Safi RI, Odde S, Shabaik Y, and Neamati N (2012). Small-molecule inhibitors of APE1 DNA repair function: an overview. Curr Mol Pharmacol 5, 14–35. - PubMed
-
- Audzevich T, Pearce G, Breucha M, Gunal G, and Jessberger R (2013). Control of the STAT6-BCL6 antagonism by SWAP-70 determines IgE production. J Immunol 190, 4946–4955. - PubMed
-
- Bernasconi NL, Traggiai E, and Lanzavecchia A (2002). Maintenance of serological memory by polyclonal activation of human memory B cells. Science 298, 2199–2202. - PubMed
-
- Brink R, Paus D, Bourne K, Hermes JR, Gardam S, Phan TG, and Chan TD (2015). The SW(HEL) system for high-resolution analysis of in vivo antigen-specific T-dependent B cell responses. Methods Mol Biol 1291, 103–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

