Revisiting Hepatitis B Virus: Challenges of Curative Therapies
- PMID: 31375584
- PMCID: PMC6798116
- DOI: 10.1128/JVI.01032-19
Revisiting Hepatitis B Virus: Challenges of Curative Therapies
Abstract
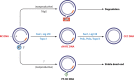
With a yearly death toll of 880,000, hepatitis B virus (HBV) remains a major health problem worldwide, despite an effective prophylactic vaccine and well-tolerated, effective antivirals. HBV causes chronic hepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma. The viral genome persists in infected hepatocytes even after long-term antiviral therapy, and its integration, though no longer able to support viral replication, destabilizes the host genome. HBV is a DNA virus that utilizes a virus-encoded reverse transcriptase to convert an RNA intermediate, termed pregenomic RNA, into the relaxed circular DNA genome, which is subsequently converted into a covalently closed circular DNA (cccDNA) in the host cell nucleus. cccDNA is maintained in the nucleus of the infected hepatocyte as a stable minichromosome and functions as the viral transcriptional template for the production of all viral gene products, and thus, it is the molecular basis of HBV persistence. The nuclear cccDNA pool can be replenished through recycling of newly synthesized, DNA-containing HBV capsids. Licensed antivirals target the HBV reverse transcriptase activity but fail to eliminate cccDNA, which would be required to cure HBV infection. Elimination of HBV cccDNA is so far only achieved by antiviral immune responses. Thus, this review will focus on possible curative strategies aimed at eliminating or crippling the viral cccDNA. Newer insights into the HBV life cycle and host immune response provide novel, potentially curative therapeutic opportunities and targets.
Keywords: Hepadnaviridae; cccDNA; hepatitis B virus; hepatocellular carcinoma; interferons; reverse transcriptase.
Copyright © 2019 American Society for Microbiology.
Figures

References
-
- Hu J. 2016. Hepatitis B virus virology and replication, p 1–34. In Liaw Y-F, Zoulim F (ed), Hepatitis B virus in human diseases. Humana Press, Totowa, NJ.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

