Modified Vaccinia Virus Ankara Can Induce Optimal CD8+ T Cell Responses to Directly Primed Antigens Depending on Vaccine Design
- PMID: 31375596
- PMCID: PMC6803277
- DOI: 10.1128/JVI.01154-19
Modified Vaccinia Virus Ankara Can Induce Optimal CD8+ T Cell Responses to Directly Primed Antigens Depending on Vaccine Design
Abstract
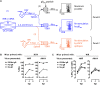
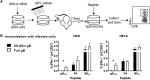
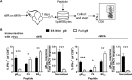
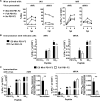
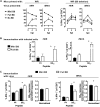
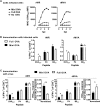
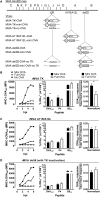
A variety of strains of vaccinia virus (VACV) have been used as recombinant vaccine vectors with the aim of inducing robust CD8+ T cell immunity. While much of the pioneering work was done with virulent strains, such as Western Reserve (WR), attenuated strains such as modified vaccinia virus Ankara (MVA) are more realistic vectors for clinical use. To unify this literature, side-by-side comparisons of virus strains are required. Here, we compare the form of antigen that supports optimal CD8+ T cell responses for VACV strains WR and MVA using equivalent constructs. We found that for multiple antigens, minimal antigenic constructs (epitope minigenes) that prime CD8+ T cells via the direct presentation pathway elicited optimal responses from both vectors, which was surprising because this finding contradicts the prevailing view in the literature for MVA. We then went on to explore the discrepancy between current and published data for MVA, finding evidence that the expression locus and in some cases the presence of the viral thymidine kinase may influence the ability of this strain to prime optimal responses from antigens that require direct presentation. This extends our knowledge of the design parameters for VACV vectored vaccines, especially those based on MVA.IMPORTANCE Recombinant vaccines based on vaccinia virus and particularly attenuated strains such as MVA are in human clinical trials, but due to the complexity of these large vectors much remains to be understood about the design parameters that alter their immunogenicity. Previous work had found that MVA vectors should be designed to express stable protein in order to induce robust immunity by CD8+ (cytotoxic) T cells. Here, we found that the primacy of stable antigen is not generalizable to all designs of MVA and may depend where a foreign antigen is inserted into the MVA genome. This unexpected finding suggests that there is an interaction between genome location and the best form of antigen for optimal T cell priming in MVA and thus possibly other vaccine vectors. It also highlights that our understanding of antigen presentation by even the best studied of vaccine vectors remains incomplete.
Keywords: CD8+ T cells; CTL; MVA; antigen presentation; antigen processing; cytotoxic T cells; live vector vaccines; modified vaccinia virus Ankara; vaccinia virus.
Copyright © 2019 Wong et al.
Figures
References
-
- Qiu J, Peng S, Ma Y, Yang A, Farmer E, Cheng MA, Roden RBS, Wu TC, Chang Y-N, Hung C-F. 2018. Epithelial boost enhances antigen expression by vaccinia virus for the generation of potent CD8+ T cell-mediated antitumor immunity following DNA priming vaccination. Virology 525:205–215. doi: 10.1016/j.virol.2018.09.019. - DOI - PMC - PubMed
-
- Marin-Lopez A, Calvo-Pinilla E, Barriales D, Lorenzo G, Brun A, Anguita J, Ortego J. 2018. CD8 T cell responses to an immunodominant epitope within the nonstructural protein NS1 provide wide immunoprotection against bluetongue virus in IFNAR–/– mice. J Virol 92. doi: 10.1128/JVI.00938-18. - DOI - PMC - PubMed
-
- Asbach B, Kibler KV, Köstler J, Perdiguero B, Yates NL, Stanfield-Oakley S, Tomaras GD, Kao S-F, Foulds KE, Roederer M, Seaman MS, Montefiori DC, Parks R, Ferrari G, Forthal DN, Phogat S, Tartaglia J, Barnett SW, Self SG, Gottardo R, Cristillo AD, Weiss DE, Galmin L, Ding S, Heeney JL, Esteban M, Jacobs BL, Pantaleo G, Wagner R. 2018. Priming with a potent HIV-1 DNA vaccine frames the quality of immune responses prior to a poxvirus and protein boost. J Virol 93:e01529-18. doi: 10.1128/JVI.01529-18. - DOI - PMC - PubMed
-
- Marlin R, Nugeyre M-T, Tchitchek N, Parenti M, Hocini H, Benjelloun F, Cannou C, Dereuddre-Bosquet N, Levy Y, Barré-Sinoussi F, Scarlatti G, Le Grand R, Menu E. 2017. Modified vaccinia virus Ankara vector induces specific cellular and humoral responses in the female reproductive tract, the main HIV portal of entry. J Immunol 199:1923–1932. doi: 10.4049/jimmunol.1700320. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

